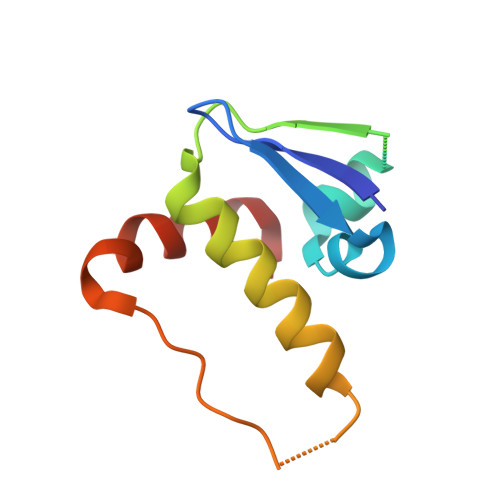
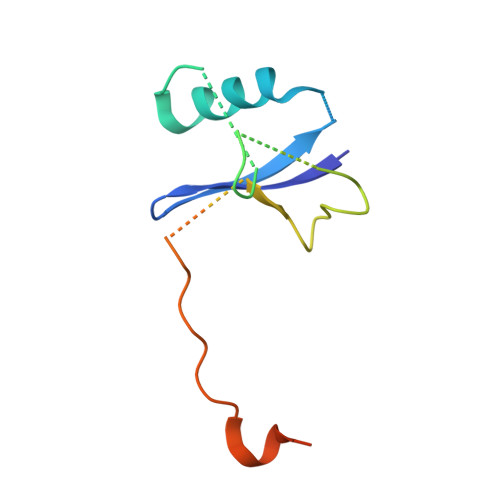
Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
Muniz, J.R.C., Guo, K., Kershaw, N.J., Ayinampudi, V., von Delft, F., Babon, J.J., Bullock, A.N.(2013) J Mol Biology 425: 3166
- PubMed: 23806657
- DOI: https://doi.org/10.1016/j.jmb.2013.06.015
- Primary Citation of Related Structures:
2WZK, 3ZKJ - PubMed Abstract:
Multi-subunit Cullin-RING E3 ligases often use repeat domain proteins as substrate-specific adaptors. Structures of these macromolecular assemblies are determined for the F-box-containing leucine-rich repeat and WD40 repeat families, but not for the suppressor of cytokine signaling (SOCS)-box-containing ankyrin repeat proteins (ASB1-18), which assemble with Elongins B and C and Cul5. We determined the crystal structures of the ternary complex of ASB9-Elongin B/C as well as the interacting N-terminal domain of Cul5 and used structural comparisons to establish a model for the complete Cul5-based E3 ligase. The structures reveal a distinct architecture of the ASB9 complex that positions the ankyrin domain coaxial to the SOCS box-Elongin B/C complex and perpendicular to other repeat protein complexes. This alternative architecture appears favorable to present the ankyrin domain substrate-binding site to the E2-ubiquitin, while also providing spacing suitable for bulky ASB9 substrates, such as the creatine kinases. The presented Cul5 structure also differs from previous models and deviates from other Cullins via a rigid-body rotation between Cullin repeats. This work highlights the adaptability of repeat domain proteins as scaffolds in substrate recognition and lays the foundation for future structure-function studies of this important E3 family.
- Structural Genomics Consortium, University of Oxford, Old Road Campus Research Building, Roosevelt Drive, Oxford OX3 7DQ, United Kingdom.
Organizational Affiliation: