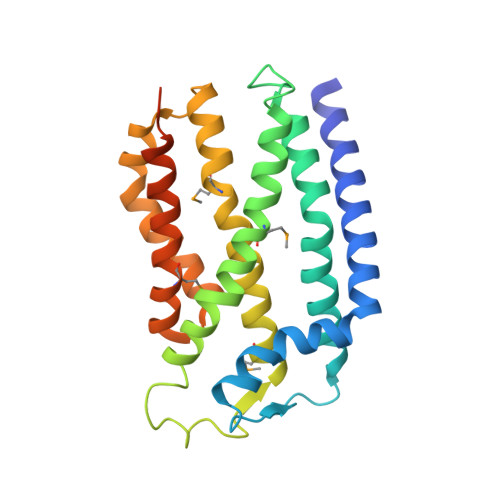
Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Rollauer, S.E., Tarry, M.J., Graham, J.E., Jaaskelainen, M., Jager, F., Johnson, S., Krehenbrink, M., Liu, S., Lukey, M.J., Marcoux, J., Mcdowell, M.A., Rodriguez, F., Roversi, P., Stansfeld, P.J., Robinson, C.V., Sansom, M.S.P., Palmer, T., Hogbom, M., Berks, B.C., Lea, S.M.(2012) Nature 49: 210
- PubMed: 23201679
- DOI: https://doi.org/10.1038/nature11683
- Primary Citation of Related Structures:
4B4A - PubMed Abstract:
The twin-arginine translocation (Tat) pathway is one of two general protein transport systems found in the prokaryotic cytoplasmic membrane and is conserved in the thylakoid membrane of plant chloroplasts. The defining, and highly unusual, property of the Tat pathway is that it transports folded proteins, a task that must be achieved without allowing appreciable ion leakage across the membrane. The integral membrane TatC protein is the central component of the Tat pathway. TatC captures substrate proteins by binding their signal peptides. TatC then recruits TatA family proteins to form the active translocation complex. Here we report the crystal structure of TatC from the hyperthermophilic bacterium Aquifex aeolicus. This structure provides a molecular description of the core of the Tat translocation system and a framework for understanding the unique Tat transport mechanism.
- Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, UK.
Organizational Affiliation: