Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
Zebisch, M., Baqi, Y., Schafer, P., Muller, C.E., Strater, N.(2014) J Struct Biol 185: 336
- PubMed: 24462745
- DOI: https://doi.org/10.1016/j.jsb.2014.01.005
- Primary Citation of Related Structures:
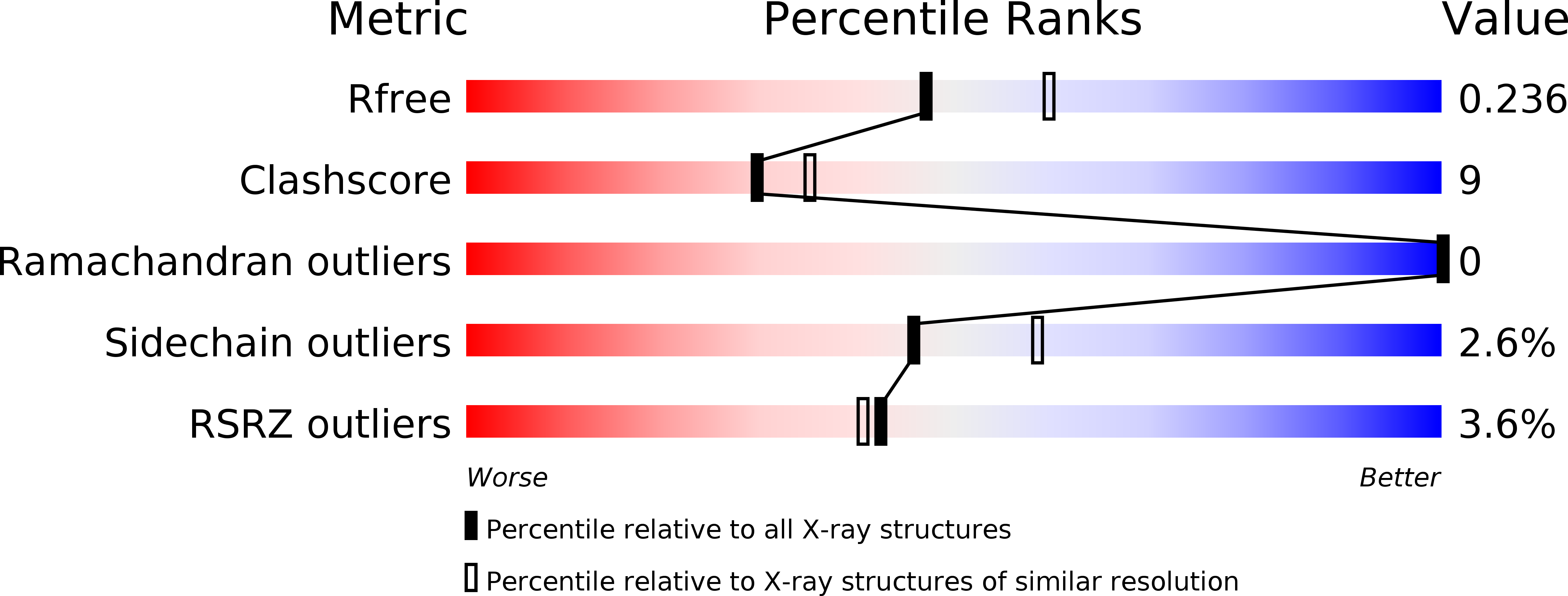
4CD1, 4CD3 - PubMed Abstract:
In many vertebrate tissues CD39-like ecto-nucleoside triphosphate diphosphohydrolases (NTPDases) act in concert with ecto-5'-nucleotidase (e5NT, CD73) to convert extracellular ATP to adenosine. Extracellular ATP is a cytotoxic, pro-inflammatory signalling molecule whereas its product adenosine constitutes a universal and potent immune suppressor. Interference with these ectonucleotidases by use of small molecule inhibitors or inhibitory antibodies appears to be an effective strategy to enhance anti-tumour immunity and suppress neoangiogenesis. Here we present the first crystal structures of an NTPDase catalytic ectodomain in complex with the Reactive Blue 2 (RB2)-derived inhibitor PSB-071. In both of the two crystal forms presented the inhibitor binds as a sandwich of two molecules at the nucleoside binding site. One of the molecules is well defined in its orientation. Specific hydrogen bonds are formed between the sulfonyl group and the nucleoside binding loop. The methylphenyl side chain functionality that improved NTPDase2-specificity is sandwiched between R245 and R394, the latter of which is exclusively found in NTPDase2. The second molecule exhibits great in-plane rotational freedom and could not be modelled in a specific orientation. In addition to this structural insight into NTPDase inhibition, the observation of the putative membrane interaction loop (MIL) in two different conformations related by a 10° rotation identifies the MIL as a dynamic section of NTPDases that is potentially involved in regulation of catalysis.
Organizational Affiliation:
Institute of Bioanalytical Chemistry, Center for Biotechnology and Biomedicine, University of Leipzig, Deutscher Platz 5, 04103 Leipzig, Germany; Division of Structural Biology, University of Oxford, Roosevelt Drive, Oxford OX3-7BN, United Kingdom. Electronic address: matthias@strubi.ox.ac.uk.