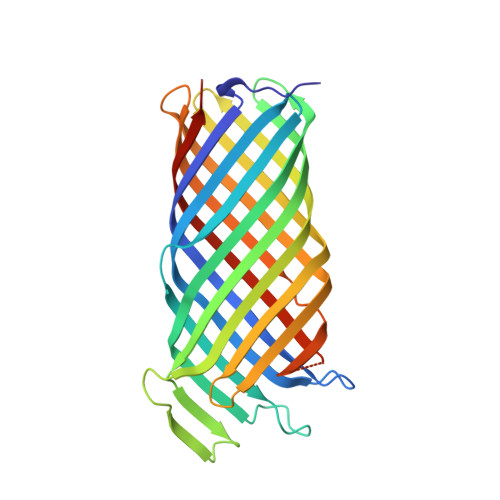
Structural basis for activation of an integral membrane protease by lipopolysaccharide.
Eren, E., van den Berg, B.(2012) J Biological Chem 287: 23971-23976
- PubMed: 22645135
- DOI: https://doi.org/10.1074/jbc.M112.376418
- Primary Citation of Related Structures:
4DCB - PubMed Abstract:
Omptins constitute a unique family of outer membrane proteases that are widespread in Enterobacteriaceae. The plasminogen activator (Pla) of Yersinia pestis is an omptin family member that is very important for development of both bubonic and pneumonic plague. The physiological function of Pla is to cleave (activate) human plasminogen to form the plasma protease plasmin. Uniquely, lipopolysaccharide (LPS) is essential for the catalytic activity of all omptins, including Pla. Why omptins require LPS for enzymatic activity is unknown. Here, we report the co-crystal structure of LPS-free Pla in complex with the activation loop peptide of human plasminogen, its natural substrate. The structure shows that in the absence of LPS, the peptide substrate binds deep within the active site groove and displaces the nucleophilic water molecule, providing an explanation for the dependence of omptins on LPS for enzymatic activity.
- Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts 01605, USA.
Organizational Affiliation: