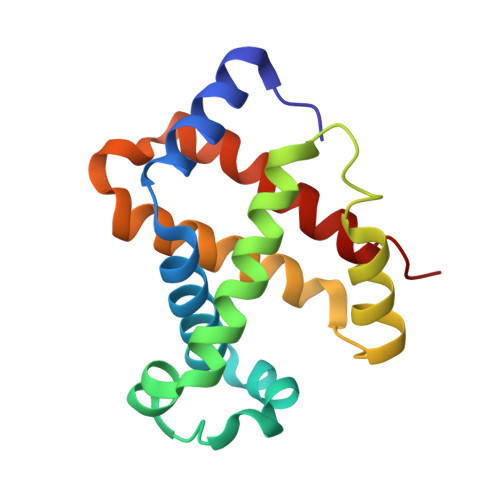
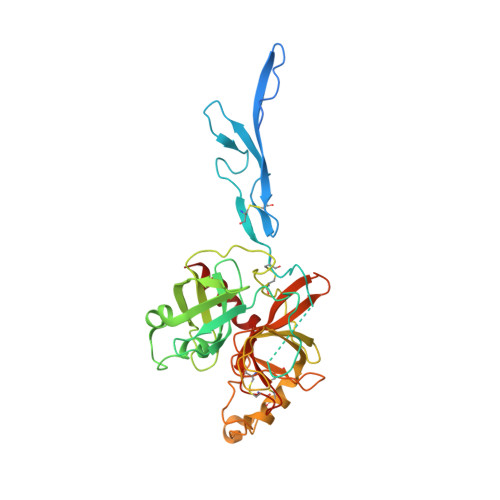
Structure of the haptoglobin-haemoglobin complex.
Andersen, C.B., Torvund-Jensen, M., Nielsen, M.J., de Oliveira, C.L., Hersleth, H.P., Andersen, N.H., Pedersen, J.S., Andersen, G.R., Moestrup, S.K.(2012) Nature 489: 456-459
- PubMed: 22922649
- DOI: https://doi.org/10.1038/nature11369
- Primary Citation of Related Structures:
4F4O - PubMed Abstract:
Red cell haemoglobin is the fundamental oxygen-transporting molecule in blood, but also a potentially tissue-damaging compound owing to its highly reactive haem groups. During intravascular haemolysis, such as in malaria and haemoglobinopathies, haemoglobin is released into the plasma, where it is captured by the protective acute-phase protein haptoglobin. This leads to formation of the haptoglobin-haemoglobin complex, which represents a virtually irreversible non-covalent protein-protein interaction. Here we present the crystal structure of the dimeric porcine haptoglobin-haemoglobin complex determined at 2.9 Å resolution. This structure reveals that haptoglobin molecules dimerize through an unexpected β-strand swap between two complement control protein (CCP) domains, defining a new fusion CCP domain structure. The haptoglobin serine protease domain forms extensive interactions with both the α- and β-subunits of haemoglobin, explaining the tight binding between haptoglobin and haemoglobin. The haemoglobin-interacting region in the αβ dimer is highly overlapping with the interface between the two αβ dimers that constitute the native haemoglobin tetramer. Several haemoglobin residues prone to oxidative modification after exposure to haem-induced reactive oxygen species are buried in the haptoglobin-haemoglobin interface, thus showing a direct protective role of haptoglobin. The haptoglobin loop previously shown to be essential for binding of haptoglobin-haemoglobin to the macrophage scavenger receptor CD163 (ref. 3) protrudes from the surface of the distal end of the complex, adjacent to the associated haemoglobin α-subunit. Small-angle X-ray scattering measurements of human haptoglobin-haemoglobin bound to the ligand-binding fragment of CD163 confirm receptor binding in this area, and show that the rigid dimeric complex can bind two receptors. Such receptor cross-linkage may facilitate scavenging and explain the increased functional affinity of multimeric haptoglobin-haemoglobin for CD163 (ref. 4).
- Department of Biomedicine, Aarhus University, 8000 Aarhus C, Denmark. cbfa@biokemi.au.dk
Organizational Affiliation: