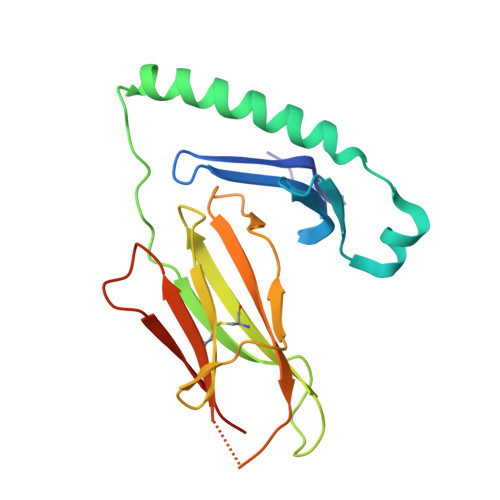
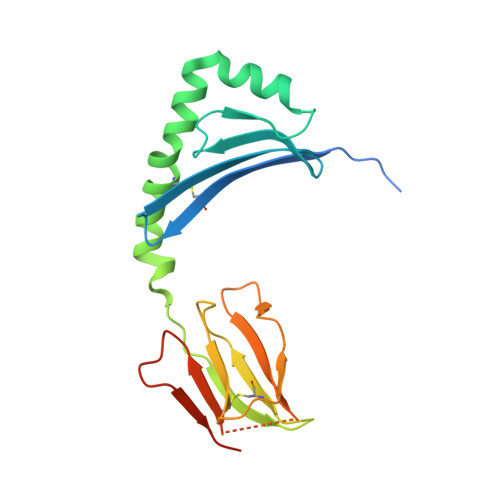
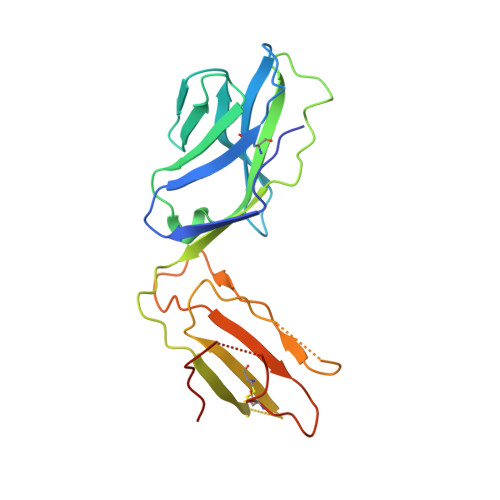
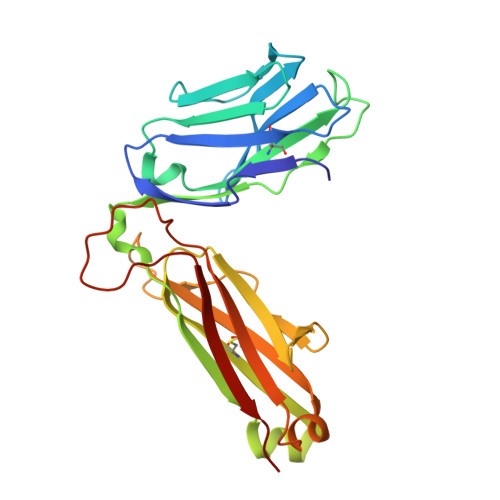
Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease.
Broughton, S.E., Petersen, J., Theodossis, A., Scally, S.W., Loh, K.L., Thompson, A., van Bergen, J., Kooy-Winkelaar, Y., Henderson, K.N., Beddoe, T., Tye-Din, J.A., Mannering, S.I., Purcell, A.W., McCluskey, J., Anderson, R.P., Koning, F., Reid, H.H., Rossjohn, J.(2012) Immunity 37: 611-621
- PubMed: 23063329
- DOI: https://doi.org/10.1016/j.immuni.2012.07.013
- Primary Citation of Related Structures:
4GG6, 4GG8 - PubMed Abstract:
Celiac disease is a human leukocyte antigen (HLA)-DQ2- and/or DQ8-associated T cell-mediated disorder that is induced by dietary gluten. Although it is established how gluten peptides bind HLA-DQ8 and HLA-DQ2, it is unclear how such peptide-HLA complexes are engaged by the T cell receptor (TCR), a recognition event that triggers disease pathology. We show that biased TCR usage (TRBV9(∗)01) underpins the recognition of HLA-DQ8-α-I-gliadin. The structure of a prototypical TRBV9(∗)01-TCR-HLA-DQ8-α-I-gliadin complex shows that the TCR docks centrally above HLA-DQ8-α-I-gliadin, in which all complementarity-determining region-β (CDRβ) loops interact with the gliadin peptide. Mutagenesis at the TRBV9(∗)01-TCR-HLA-DQ8-α-I-gliadin interface provides an energetic basis for the Vβ bias. Moreover, CDR3 diversity accounts for TRBV9(∗)01(+) TCRs exhibiting differing reactivities toward the gliadin epitopes at various deamidation states. Accordingly, biased TCR usage is an important factor in the pathogenesis of DQ8-mediated celiac disease.
- The Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: