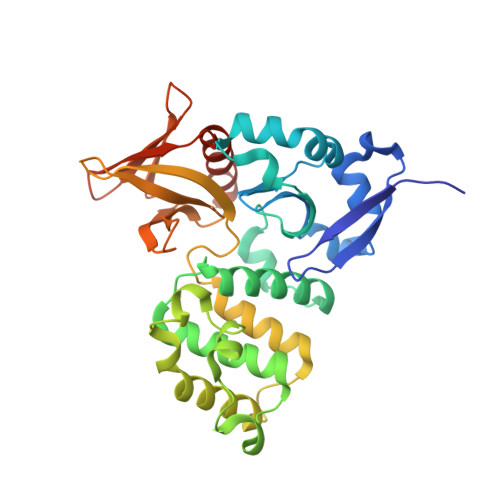
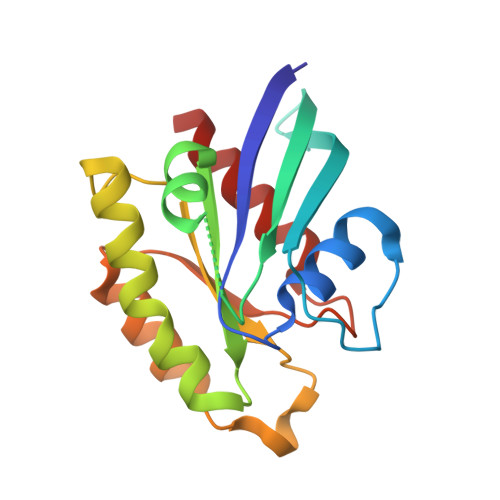
The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) Bound to Both the Rap1 GTPase and the Heart of Glass (HEG1) Cytoplasmic Tail.
Gingras, A.R., Puzon-McLaughlin, W., Ginsberg, M.H.(2013) J Biological Chem 288: 23639-23649
- PubMed: 23814056
- DOI: https://doi.org/10.1074/jbc.M113.462911
- Primary Citation of Related Structures:
4HDO, 4HDQ - PubMed Abstract:
Loss of function mutation in Krev interaction trapped 1 (KRIT1) causes autosomal dominant familial cerebral cavernous malformations and disrupts cardiovascular development. The biological function of KRIT1 requires that its FERM (band 4.1, ezrin, radixin, moesin) domain physically interact with both the small GTPase Rap1 and the cytoplasmic tail of the Heart of glass (HEG1) membrane anchor. In this study, we show that the KRIT1 FERM domain can bind both Rap1 and HEG1 simultaneously, and we solved the crystal structure of the KRIT1-Rap1-HEG1 ternary complex. Rap1 binds on the surface of the F1 and F2 subdomains, in an interaction that leaves its Switch II region accessible to other potential effectors. HEG1 binds in a hydrophobic pocket at the KRIT1 F1 and F3 interface, and there is no overlap with the Rap1-binding site. Indeed, the affinity of KRIT1 or the KRIT1-Rap1 complex for HEG1 is comparable (Kd = 1.2 and 0.96 μm, respectively) showing that there is no competition between the two sites. Furthermore, analysis of this structure revealed a specific ionic interaction between the F2 lobe of KRIT1 and Rap1 that could explain the remarkable Rap1 specificity of KRIT1. This structural insight enabled design of KRIT1(K570I), a mutant that binds Rap1 with 8-fold lower affinity and exhibits increased binding to HRas. These data show that HEG1 can recruit the Rap1-KRIT complex to the plasma membrane where Rap1's Switch II region remains accessible and reveals an important determinant of KRIT1's specificity for Rap1.
- Department of Medicine, University of California at San Diego, La Jolla, California 92093, USA. agingras@ucsd.edu
Organizational Affiliation: