Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5.
Lucas, M., Gaspar, A.H., Pallara, C., Rojas, A.L., Fernandez-Recio, J., Machner, M.P., Hierro, A.(2014) Proc Natl Acad Sci U S A 111: E3514-E3523
- PubMed: 25114243
- DOI: https://doi.org/10.1073/pnas.1405391111
- Primary Citation of Related Structures:
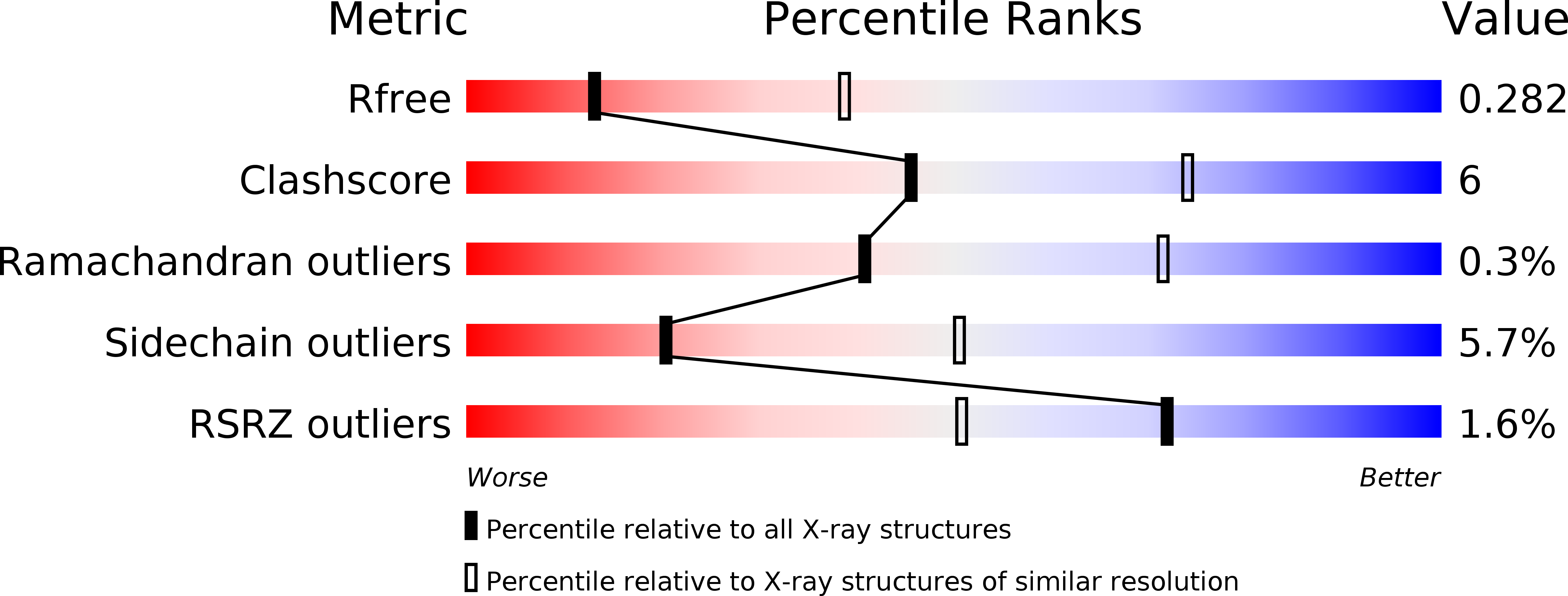
4KYI - PubMed Abstract:
A challenge for microbial pathogens is to assure that their translocated effector proteins target only the correct host cell compartment during infection. The Legionella pneumophila effector vacuolar protein sorting inhibitor protein D (VipD) localizes to early endosomal membranes and alters their lipid and protein composition, thereby protecting the pathogen from endosomal fusion. This process requires the phospholipase A1 (PLA1) activity of VipD that is triggered specifically on VipD binding to the host cell GTPase Rab5, a key regulator of endosomes. Here, we present the crystal structure of VipD in complex with constitutively active Rab5 and reveal the molecular mechanism underlying PLA1 activation. An active site-obstructing loop that originates from the C-terminal domain of VipD is repositioned on Rab5 binding, thereby exposing the catalytic pocket within the N-terminal PLA1 domain. Substitution of amino acid residues located within the VipD-Rab5 interface prevented Rab5 binding and PLA1 activation and caused a failure of VipD mutant proteins to target to Rab5-enriched endosomal structures within cells. Experimental and computational analyses confirmed an extended VipD-binding interface on Rab5, explaining why this L. pneumophila effector can compete with cellular ligands for Rab5 binding. Together, our data explain how the catalytic activity of a microbial effector can be precisely linked to its subcellular localization.
Organizational Affiliation:
Structural Biology Unit, Center for Cooperative Research in Biosciences, 48160 Derio, Spain;