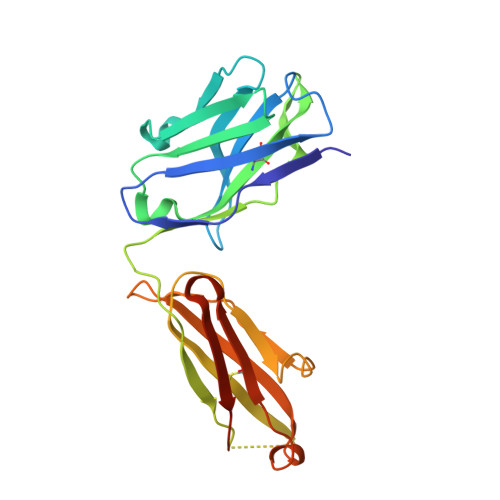
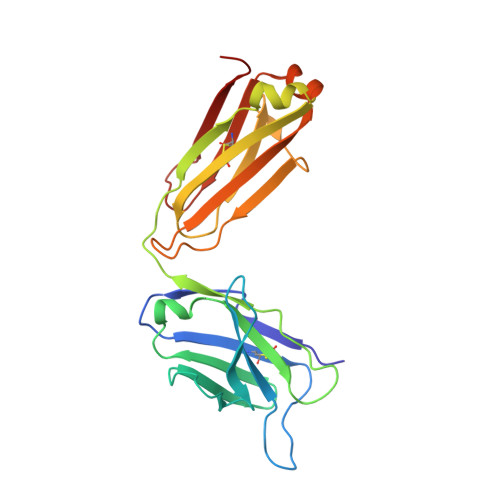
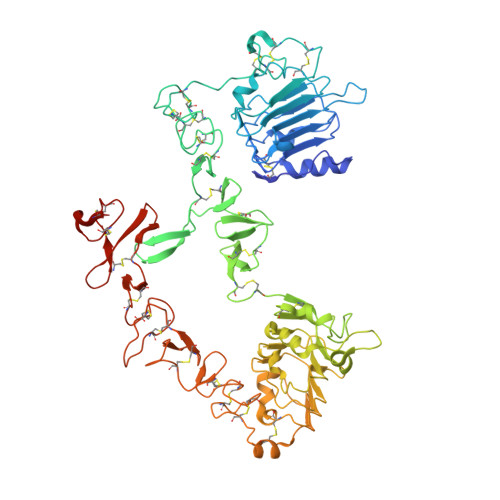
RG7116, a Therapeutic Antibody That Binds the Inactive HER3 Receptor and Is Optimized for Immune Effector Activation.
Mirschberger, C., Schiller, C.B., Schraml, M., Dimoudis, N., Friess, T., Gerdes, C.A., Reiff, U., Lifke, V., Hoelzlwimmer, G., Kolm, I., Hopfner, K.P., Niederfellner, G., Bossenmaier, B.(2013) Cancer Res 73: 5183-5194
- PubMed: 23780344
- DOI: https://doi.org/10.1158/0008-5472.CAN-13-0099
- Primary Citation of Related Structures:
4LEO - PubMed Abstract:
The EGF receptor (EGFR) HER3 is emerging as an attractive cancer therapeutic target due to its central position in the HER receptor signaling network. HER3 amplifies phosphoinositide 3-kinase (PI3K)-driven tumorigenesis and its upregulation in response to other anti-HER therapies has been implicated in resistance to them. Here, we report the development and characterization of RG7116, a novel anti-HER3 monoclonal antibody (mAb) designed to block HER3 activation, downregulate HER3, and mediate enhanced antibody-dependent cell-mediated cytotoxicity (ADCC) via glycoengineering of the Fc moiety. Biochemical studies and X-ray crystallography revealed that RG7116 bound potently and selectively to domain 1 of human HER3. Heregulin binding was prevented by RG7116 at concentrations more than 1 nmol/L as was nearly complete inhibition of HER3 heterodimerization and phosphorylation, thereby preventing downstream AKT phosphorylation. In vivo RG7116 treatment inhibited xenograft tumor growth up to 90% relative to controls in a manner accompanied by downregulation of cell surface HER3. RG7116 efficacy was further enhanced in combination with anti-EGFR (RG7160) or anti-HER2 (pertuzumab) mAbs. Furthermore, the ADCC potency of RG7116 was enhanced compared with the nonglycoengineered parental antibody, both in vitro and in orthotopic tumor xenograft models, where an increased median survival was documented. ADCC degree achieved in vitro correlated with HER3 expression levels on tumor cells. In summary, the combination of strong signaling inhibition and enhanced ADCC capability rendered RG7116 a highly potent HER3-targeting agent suitable for clinical development.
- Pharma Research and Early Development (pRED), Roche Diagnostics GmbH, Penzberg, Germany. brigit.bossenmaier@roche.com
Organizational Affiliation: