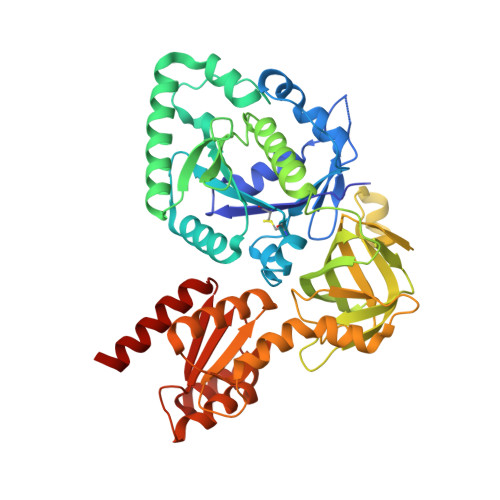
eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining.
Kuhle, B., Ficner, R.(2014) EMBO J 33: 1177-1191
- PubMed: 24686316
- DOI: https://doi.org/10.1002/embj.201387344
- Primary Citation of Related Structures:
4N3G, 4N3N, 4N3S, 4NCF, 4NCL, 4NCN - PubMed Abstract:
eIF5B is a eukaryal translational GTPase that catalyzes ribosomal subunit joining to form elongation-competent ribosomes. Despite its central role in protein synthesis, the mechanistic details that govern the function of eIF5B or its archaeal and bacterial (IF2) orthologs remained unclear. Here, we present six high-resolution crystal structures of eIF5B in its apo, GDP- and GTP-bound form that, together with an analysis of the thermodynamics of nucleotide binding, provide a detailed picture of the entire nucleotide cycle performed by eIF5B. Our data show that GTP binding induces significant conformational changes in the two conserved switch regions of the G domain, resulting in the reorganization of the GTPase center. These rearrangements are accompanied by the rotation of domain II relative to the G domain and release of domain III from its stable contacts with switch 2, causing an increased intrinsic flexibility in the free GTP-bound eIF5B. Based on these data, we propose a novel domain release mechanism for eIF5B/IF2 activation that explains how eIF5B and IF2 fulfill their catalytic role during ribosomal subunit joining.
- Abteilung für Molekulare Strukturbiologie, Institut für Mikrobiologie und Genetik Göttinger Zentrum für Molekulare Biowissenschaften Georg-August-Universität Göttingen, Göttingen, Germany bkuhle@gwdg.de.
Organizational Affiliation: