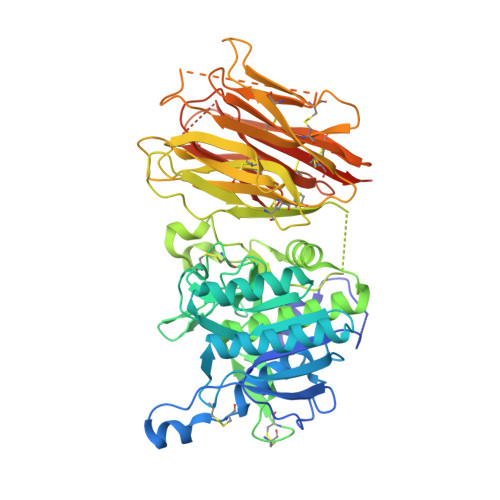
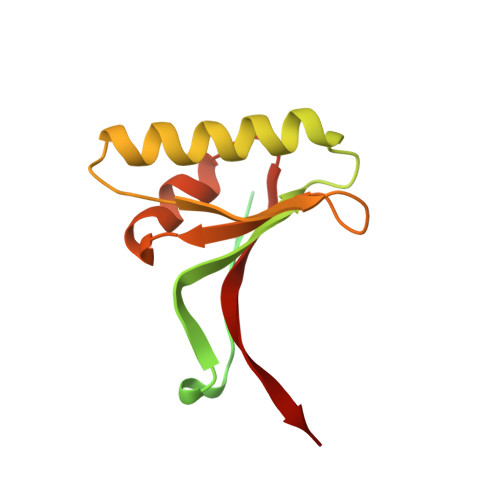
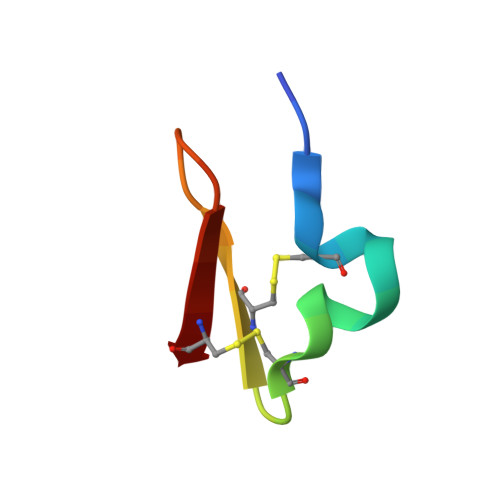
Design and synthesis of truncated EGF-A peptides that restore LDL-R recycling in the presence of PCSK9 in vitro.
Schroeder, C.I., Swedberg, J.E., Withka, J.M., Rosengren, K.J., Akcan, M., Clayton, D.J., Daly, N.L., Cheneval, O., Borzilleri, K.A., Griffor, M., Stock, I., Colless, B., Walsh, P., Sunderland, P., Reyes, A., Dullea, R., Ammirati, M., Liu, S., McClure, K.F., Tu, M., Bhattacharya, S.K., Liras, S., Price, D.A., Craik, D.J.(2014) Chem Biol 21: 284-294
- PubMed: 24440079
- DOI: https://doi.org/10.1016/j.chembiol.2013.11.014
- Primary Citation of Related Structures:
2MG9, 4NE9 - PubMed Abstract:
Disrupting the binding interaction between proprotein convertase (PCSK9) and the epidermal growth factor-like domain A (EGF-A domain) in the low-density lipoprotein receptor (LDL-R) is a promising strategy to promote LDL-R recycling and thereby lower circulating cholesterol levels. In this study, truncated 26 amino acid EGF-A analogs were designed and synthesized, and their structures were analyzed in solution and in complex with PCSK9. The most potent peptide had an increased binding affinity for PCSK9 (KD = 0.6 μM) compared with wild-type EGF-A (KD = 1.2 μM), and the ability to increase LDL-R recycling in the presence of PCSK9 in a cell-based assay.
- Institute for Molecular Bioscience, The University of Queensland, Brisbane, 4072 QLD, Australia.
Organizational Affiliation: