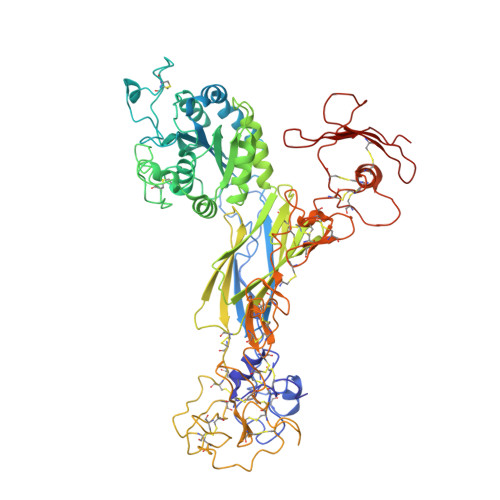
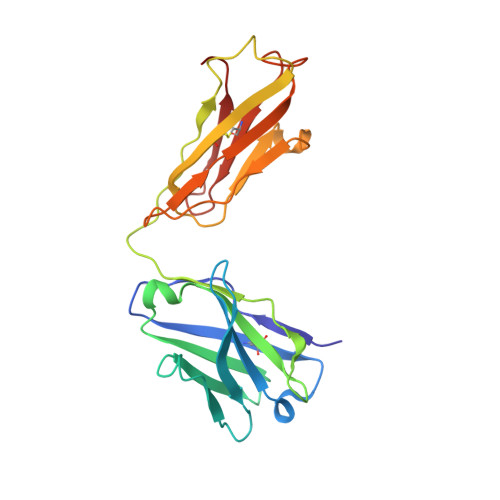
Atomic basis for the species-specific inhibition of alpha V integrins by monoclonal antibody 17E6 is revealed by the crystal structure of alpha V beta 3 ectodomain-17E6 Fab complex.
Mahalingam, B., Van Agthoven, J.F., Xiong, J.P., Alonso, J.L., Adair, B.D., Rui, X., Anand, S., Mehrbod, M., Mofrad, M.R., Burger, C., Goodman, S.L., Arnaout, M.A.(2014) J Biological Chem 289: 13801-13809
- PubMed: 24692540
- DOI: https://doi.org/10.1074/jbc.M113.546929
- Primary Citation of Related Structures:
4O02 - PubMed Abstract:
The function-blocking, non-RGD-containing, and primate-specific mouse monoclonal antibody 17E6 binds the αV subfamily of integrins. 17E6 is currently in phase II clinical trials for treating cancer. To elucidate the structural basis of recognition and the molecular mechanism of inhibition, we crystallized αVβ3 ectodomain in complex with the Fab fragment of 17E6. Protein crystals grew in presence of the activating cation Mn(2+). The integrin in the complex and in solution assumed the genuflected conformation. 17E6 Fab bound exclusively to the Propeller domain of the αV subunit. At the core of αV-Fab interface were interactions involving Propeller residues Lys-203 and Gln-145, with the latter accounting for primate specificity. The Propeller residue Asp-150, which normally coordinates Arg of the ligand Arg-Gly-Asp motif, formed contacts with Arg-54 of the Fab that were expected to reduce soluble FN10 binding to cellular αVβ3 complexed with 17E6. This was confirmed in direct binding studies, suggesting that 17E6 is an allosteric inhibitor of αV integrins.
- From the Structural Biology Program and.
Organizational Affiliation: