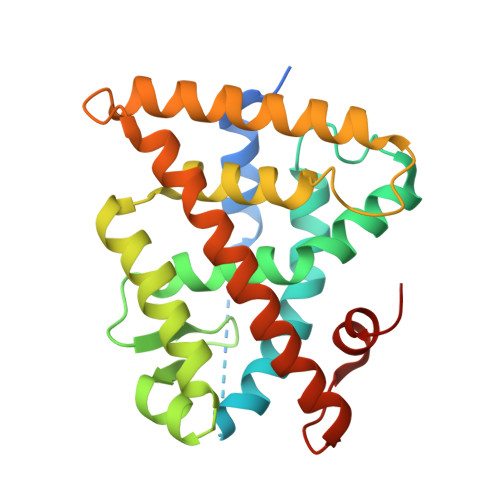
A natural-product switch for a dynamic protein interface.
Scheepstra, M., Nieto, L., Hirsch, A.K., Fuchs, S., Leysen, S., Lam, C.V., in het Panhuis, L., van Boeckel, C.A., Wienk, H., Boelens, R., Ottmann, C., Milroy, L.G., Brunsveld, L.(2014) Angew Chem Int Ed Engl 53: 6443-6448
- PubMed: 24821627
- DOI: https://doi.org/10.1002/anie.201403773
- Primary Citation of Related Structures:
4OC7 - PubMed Abstract:
Small ligands are a powerful way to control the function of protein complexes via dynamic binding interfaces. The classic example is found in gene transcription where small ligands regulate nuclear receptor binding to coactivator proteins via the dynamic activation function 2 (AF2) interface. Current ligands target the ligand-binding pocket side of the AF2. Few ligands are known, which selectively target the coactivator side of the AF2, or which can be selectively switched from one side of the interface to the other. We use NMR spectroscopy and modeling to identify a natural product, which targets the retinoid X receptor (RXR) at both sides of the AF2. We then use chemical synthesis, cellular screening and X-ray co-crystallography to split this dual activity, leading to a potent and molecularly efficient RXR agonist, and a first-of-kind inhibitor selective for the RXR/coactivator interaction. Our findings justify future exploration of natural products at dynamic protein interfaces.
- Laboratory of Chemical Biology and Institute of Complex Molecular Systems (ICMS), Department of Biomedical Engineering, Technische Universiteit Eindhoven, Den Dolech 2, 5612 AZ Eindhoven (The Netherlands) http://www.tue.nl/cb.
Organizational Affiliation: