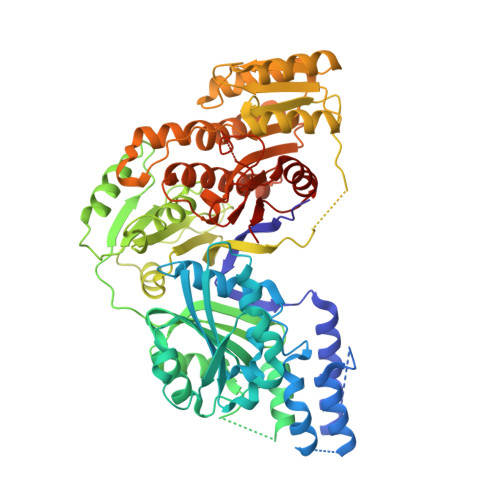
A human fatty acid synthase inhibitor binds beta-ketoacyl reductase in the keto-substrate site.
Hardwicke, M.A., Rendina, A.R., Williams, S.P., Moore, M.L., Wang, L., Krueger, J.A., Plant, R.N., Totoritis, R.D., Zhang, G., Briand, J., Burkhart, W.A., Brown, K.K., Parrish, C.A.(2014) Nat Chem Biol 10: 774-779
- PubMed: 25086508
- DOI: https://doi.org/10.1038/nchembio.1603
- Primary Citation of Related Structures:
4PIV - PubMed Abstract:
Human fatty acid synthase (hFAS) is a complex, multifunctional enzyme that is solely responsible for the de novo synthesis of long chain fatty acids. hFAS is highly expressed in a number of cancers, with low expression observed in most normal tissues. Although normal tissues tend to obtain fatty acids from the diet, tumor tissues rely on de novo fatty acid synthesis, making hFAS an attractive metabolic target for the treatment of cancer. We describe here the identification of GSK2194069, a potent and specific inhibitor of the β-ketoacyl reductase (KR) activity of hFAS; the characterization of its enzymatic and cellular mechanism of action; and its inhibition of human tumor cell growth. We also present the design of a new protein construct suitable for crystallography, which resulted in what is to our knowledge the first co-crystal structure of the human KR domain and includes a bound inhibitor.
- 1] Oncology Research and Development, GlaxoSmithKline, Collegeville, Pennsylvania, USA. [2].
Organizational Affiliation: