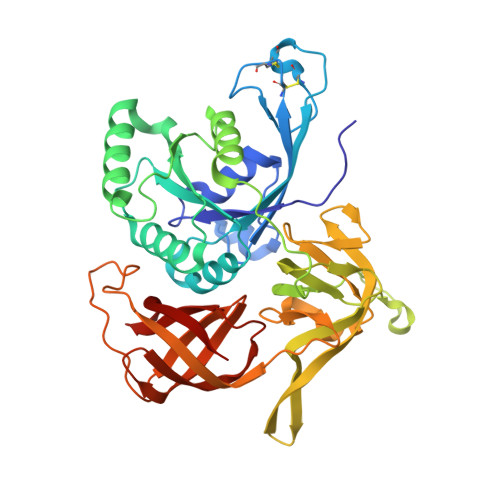
Crystal structure of gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDPCP at 1.64A resolution
Kravchenko, O.V., Nikonov, O.S., Arhipova, V.I., Stolboushkina, E.A., Gabdulkhakov, A.G., Nikulin, A.D., Garber, M.B., Nikonov, S.V.To be published.