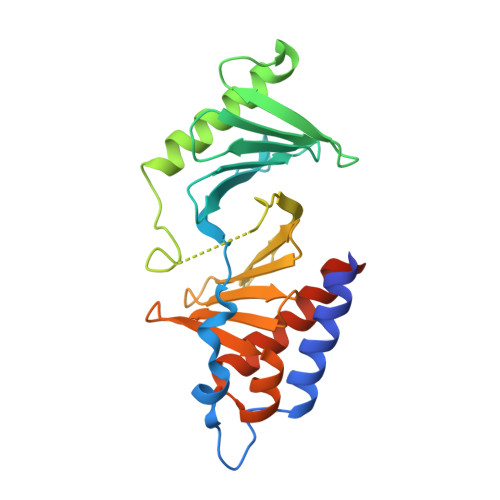
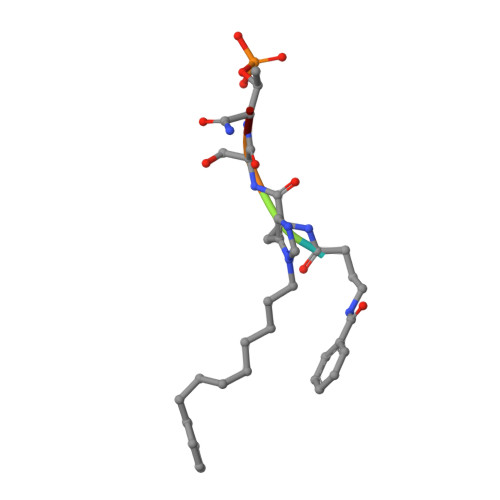
A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
Ahn, M., Han, Y.H., Park, J.E., Kim, S., Lee, W.C., Lee, S.J., Gunasekaran, P., Cheong, C., Shin, S.Y., Kim, H.Y., Ryu, E.K., Murugan, R.N., Kim, N.H., Bang, J.K.(2015) J Med Chem 58: 294-304
- PubMed: 25347203
- DOI: https://doi.org/10.1021/jm501147g
- Primary Citation of Related Structures:
4RCP, 4WHH, 4WHK, 4WHL - PubMed Abstract:
Recent progress in the development of peptide-derived Polo-like kinase (Plk1) polo-box domain (PBD) inhibitors has led to the synthesis of multiple peptide ligands with high binding affinity and selectivity. However, few systematic analyses have been conducted to identify key Plk1 residues and characterize their interactions with potent Plk1 peptide inhibitors. We performed systematic deletion analysis using the most potent 4j peptide and studied N-terminal capping of the minimal peptide with diverse organic moieties, leading to the identification of the peptidomimetic 8 (AB-103) series with high binding affinity and selectivity. To evaluate the bioavailability of short peptidomimetic ligands, PEGylated 8 series were synthesized and incubated with HeLa cells to test for cellular uptake, antiproliferative activity, and Plk1 kinase inhibition. Finally, crystallographic studies of the Plk1 PBD in complex with peptidomimetics 8 and 22 (AB-103-5) revealed the presence of two hydrogen bond interactions responsible for their high binding affinity and selectivity.
- Division of Magnetic Resonance, Korea Basic Science Institute , 804-1, Yangcheong Ri, Ochang, Chungbuk, Cheongwon 363-883, Republic of Korea.
Organizational Affiliation: