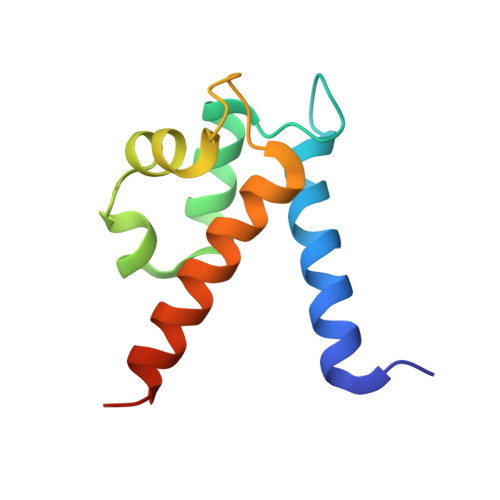
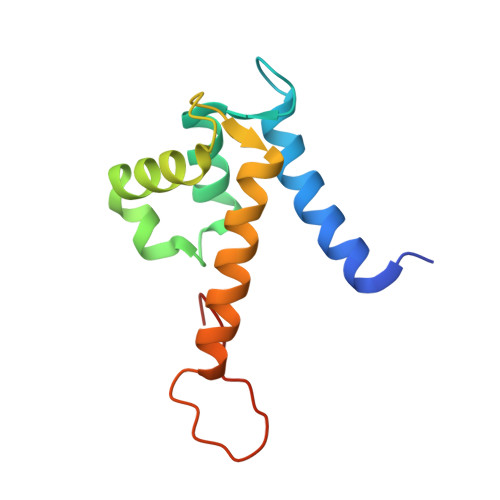
Manganese Binding Properties of Human Calprotectin under Conditions of High and Low Calcium: X-ray Crystallographic and Advanced Electron Paramagnetic Resonance Spectroscopic Analysis.
Gagnon, D.M., Brophy, M.B., Bowman, S.E., Stich, T.A., Drennan, C.L., Britt, R.D., Nolan, E.M.(2015) J Am Chem Soc 137: 3004-3016
- PubMed: 25597447
- DOI: https://doi.org/10.1021/ja512204s
- Primary Citation of Related Structures:
4XJK - PubMed Abstract:
The antimicrobial protein calprotectin (CP), a hetero-oligomer of the S100 family members S100A8 and S100A9, is the only identified mammalian Mn(II)-sequestering protein. Human CP uses Ca(II) ions to tune its Mn(II) affinity at a biologically unprecedented hexahistidine site that forms at the S100A8/S100A9 interface, and the molecular basis for this phenomenon requires elucidation. Herein, we investigate the remarkable Mn(II) coordination chemistry of human CP using X-ray crystallography as well as continuous-wave (CW) and pulse electron paramagnetic resonance (EPR) spectroscopies. An X-ray crystallographic structure of Mn(II)-CP containing one Mn(II), two Ca(II), and two Na(I) ions per CP heterodimer is reported. The CW EPR spectrum of Ca(II)- and Mn(II)-bound CP prepared with a 10:0.9:1 Ca(II):Mn(II):CP ratio is characterized by an unusually low zero-field splitting of 485 MHz (E/D = 0.30) for the S = 5/2 Mn(II) ion, consistent with the high symmetry of the His6 binding site observed crystallographically. Results from electron spin-echo envelope modulation and electron-nuclear double resonance experiments reveal that the six Mn(II)-coordinating histidine residues of Ca(II)- and Mn(II)-bound CP are spectroscopically equivalent. The observed (15)N (I = 1/2) hyperfine couplings (A) arise from two distinct classes of nitrogen atoms: the coordinating ε-nitrogen of the imidazole ring of each histidine ligand (A = [3.45, 3.71, 5.91] MHz) and the distal δ-nitrogen (A = [0.11, 0.18, 0.42] MHz). In the absence of Ca(II), the binding affinity of CP for Mn(II) drops by two to three orders of magnitude and coincides with Mn(II) binding at the His6 site as well as other sites. This study demonstrates the role of Ca(II) in enabling high-affinity and specific binding of Mn(II) to the His6 site of human calprotectin.
- Department of Chemistry, University of California , Davis, California 95616, United States.
Organizational Affiliation: