Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
Blanchetot, C., De Jonge, N., Desmyter, A., Ongenae, N., Hofman, E., Klarenbeek, A., Sadi, A., Hultberg, A., Kretz-Rommel, A., Spinelli, S., Loris, R., Cambillau, C., de Haard, H.(2016) J Biological Chem 291: 13846-13854
- PubMed: 27129274
- DOI: https://doi.org/10.1074/jbc.M115.695528
- Primary Citation of Related Structures:
4ZS7 - PubMed Abstract:
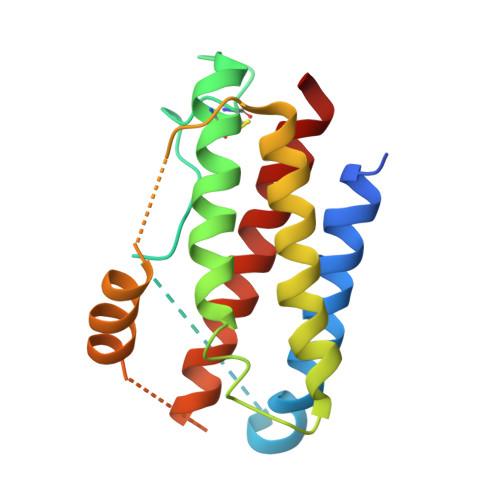
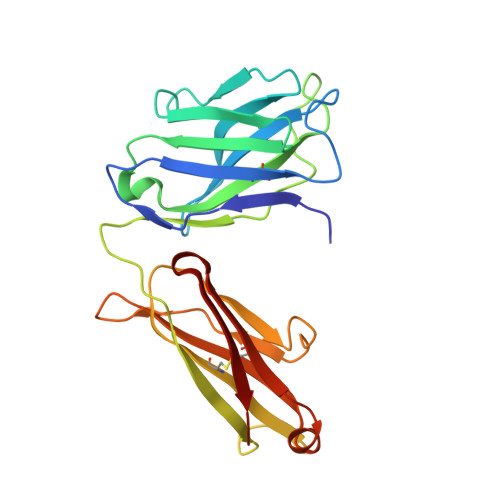
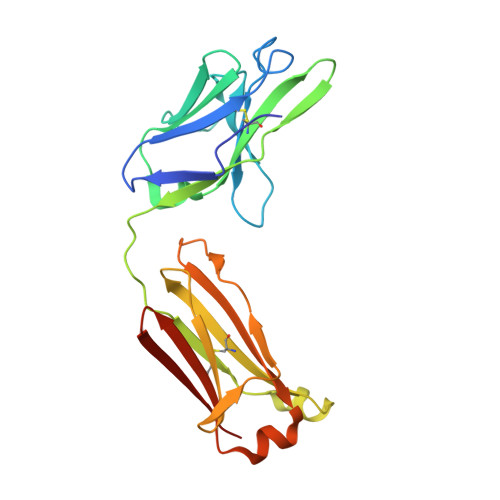
Interleukin 6 plays a key role in mediating inflammatory reactions in autoimmune diseases and cancer, where it is also involved in metastasis and tissue invasion. Neutralizing antibodies against IL-6 and its receptor have been approved for therapeutic intervention or are in advanced stages of clinical development. Here we describe the crystal structures of the complexes of IL-6 with two Fabs derived from conventional camelid antibodies that antagonize the interaction between the cytokine and its receptor. The x-ray structures of these complexes provide insights into the mechanism of neutralization by the two antibodies and explain the very high potency of one of the antibodies. It effectively competes for binding to the cytokine with IL-6 receptor (IL-6R) by using side chains of two CDR residues filling the site I cavities of IL-6, thus mimicking the interactions of Phe(229) and Phe(279) of IL-6R. In the first antibody, a HCDR3 tryptophan binds similarly to hot spot residue Phe(279) Mutation of this HCDR3 Trp residue into any other residue except Tyr or Phe significantly weakens binding of the antibody to IL-6, as was also observed for IL-6R mutants of Phe(279) In the second antibody, the side chain of HCDR3 valine ties into site I like IL-6R Phe(279), whereas a LCDR1 tyrosine side chain occupies a second cavity within site I and mimics the interactions of IL-6R Phe(229).
- From argenx, 9052 Zwijnaarde, Belgium.
Organizational Affiliation: