9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice.
Ruhl, R., Krzyzosiak, A., Niewiadomska-Cimicka, A., Rochel, N., Szeles, L., Vaz, B., Wietrzych-Schindler, M., Alvarez, S., Szklenar, M., Nagy, L., de Lera, A.R., Krezel, W.(2015) PLoS Genet 11: e1005213-e1005213
- PubMed: 26030625
- DOI: https://doi.org/10.1371/journal.pgen.1005213
- Primary Citation of Related Structures:
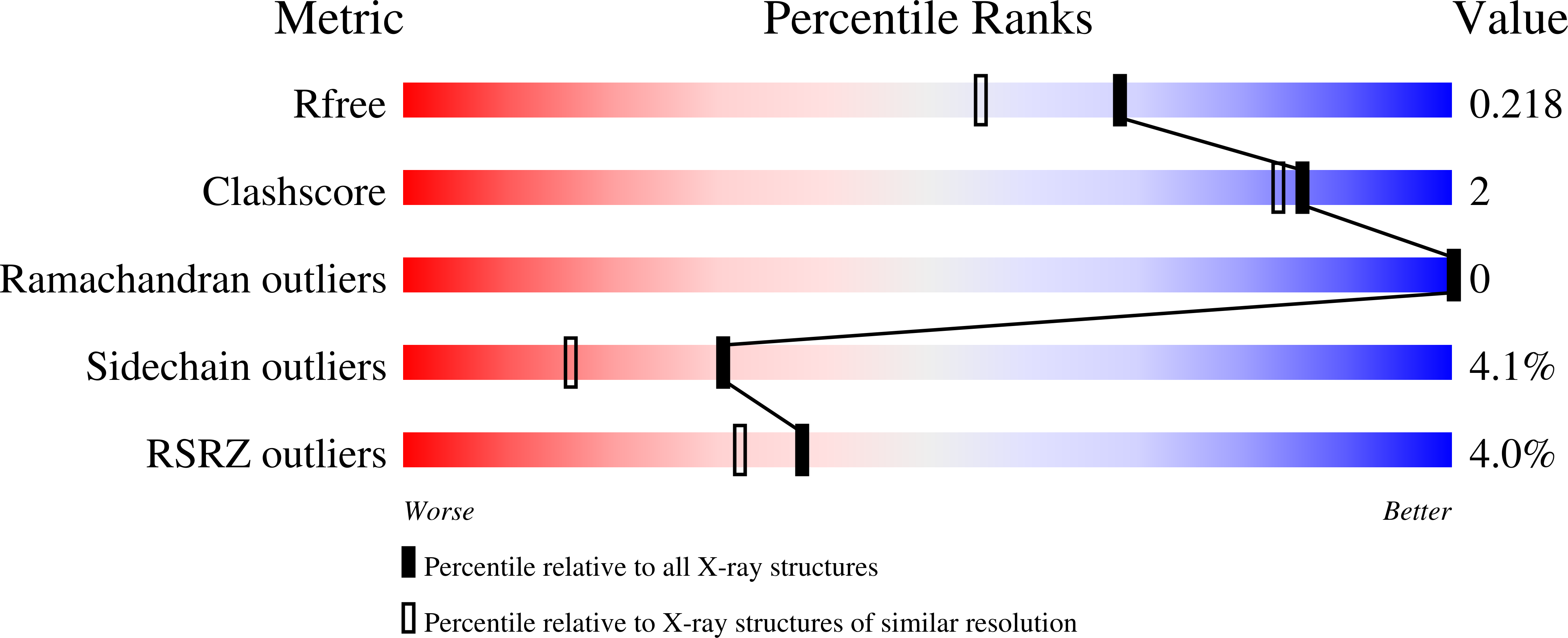
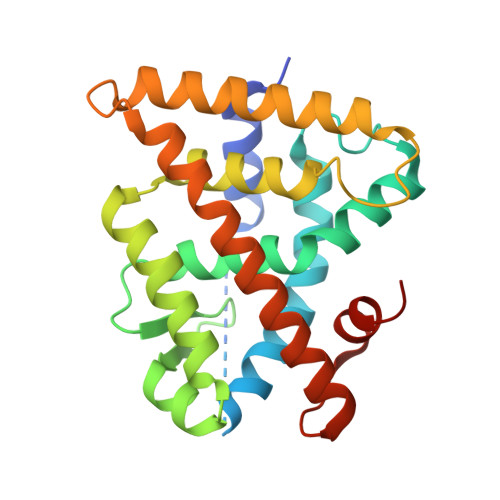
4ZSH - PubMed Abstract:
The retinoid X receptors (RXRs) are ligand-activated transcription factors which heterodimerize with a number of nuclear hormone receptors, thereby controlling a variety of (patho)-physiological processes. Although synthetic RXR ligands are developed for the treatment of various diseases, endogenous ligand(s) for these receptors have not been conclusively identified. We show here that mice lacking cellular retinol binding protein (Rbp1-/-) display memory deficits reflecting compromised RXR signaling. Using HPLC-MS and chemical synthesis we identified in Rbp1-/- mice reduced levels of 9-cis-13,14-dihydroretinoic acid (9CDHRA), which acts as an RXR ligand since it binds and transactivates RXR in various assays. 9CDHRA rescues the Rbp1-/- phenotype similarly to a synthetic RXR ligand and displays similar transcriptional activity in cultured human dendritic cells. High endogenous levels of 9CDHRA in mice indicate physiological relevance of these data and that 9CDHRA acts as an endogenous RXR ligand.
- Department of Biochemistry and Molecular Biology, Faculty of Medicine, Debrecen, Hungary; Paprika Bioanalytics BT, Debrecen, Hungary.
Organizational Affiliation: