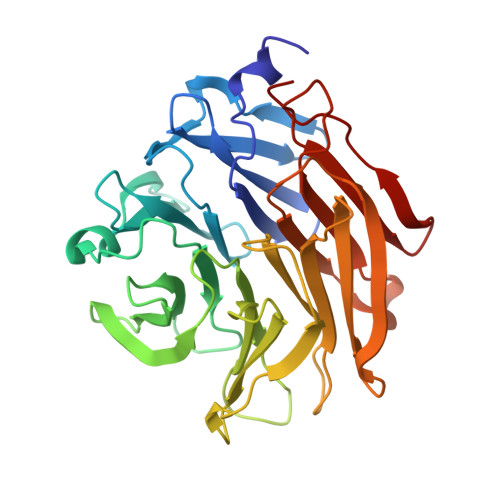
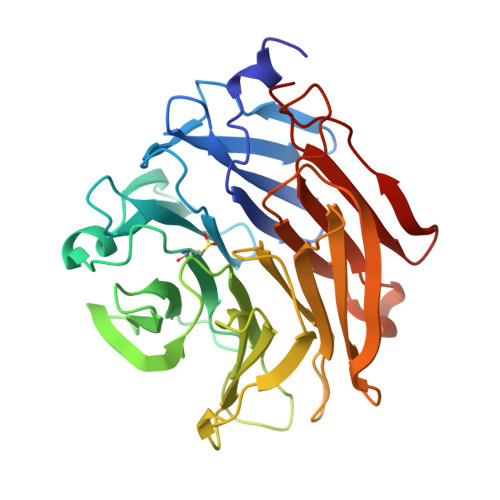
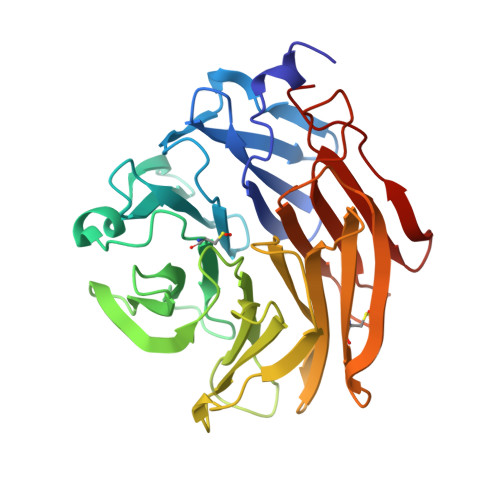
Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Houser, J., Komarek, J., Cioci, G., Varrot, A., Imberty, A., Wimmerova, M.(2015) Acta Crystallogr D Biol Crystallogr 71: 442
- PubMed: 25760594
- DOI: https://doi.org/10.1107/S1399004714026595
- Primary Citation of Related Structures:
4AGT, 4AH4, 4AHA, 4C1Y, 4D4U, 4D52, 4UOU - PubMed Abstract:
The Aspergillus fumigatus lectin AFL was recently described as a new member of the AAL lectin family. As a lectin from an opportunistic pathogen, it might play an important role in the interaction of the pathogen with the human host. A detailed study of structures of AFL complexed with several monosaccharides and oligosaccharides, including blood-group epitopes, was combined with affinity data from SPR and discussed in the context of previous findings. Its six binding sites are non-equivalent, and owing to minor differences in amino-acid composition they exhibit a marked difference in specific ligand recognition. AFL displays a high affinity in the micromolar range towards oligosaccharides which were detected in plants and also those bound on the human epithelia. All of these results indicate AFL to be a complex member of the lectin family and a challenging target for future medical research and, owing to its binding properties, a potentially useful tool in specific biotechnological applications.
- Central European Institute of Technology, Masaryk University, Kamenice 5, 62500 Brno, Czech Republic.
Organizational Affiliation: