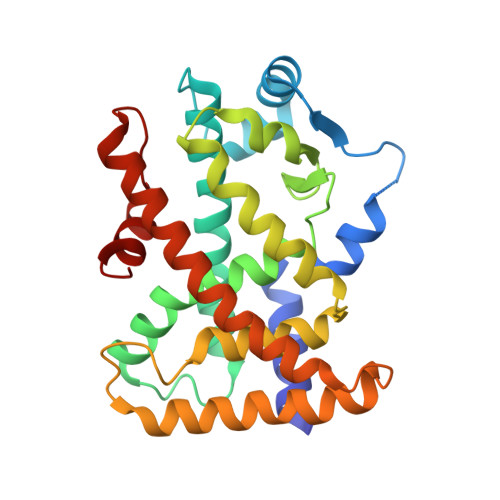
17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Egawa, D., Itoh, T., Akiyama, Y., Saito, T., Yamamoto, K.(2016) ACS Chem Biol
- PubMed: 27337155
- DOI: https://doi.org/10.1021/acschembio.6b00338
- Primary Citation of Related Structures:
5AZT, 5AZV - PubMed Abstract:
17-Hydroxy docosahexaenoic acid (17-HDHA) is an oxidized form of docosahexaenoic acid (DHA) and known as a specialized proresolving mediator. We found that a further oxidized product, 17-oxodocosahexaenoic acid (17-oxoDHA), activates peroxisome proliferator-activated receptors γ (PPARγ) and PPARα in transcriptional assays and thus can be classified as an α/γ dual agonist. ESI mass spectroscopy and X-ray crystallographic analysis showed that 17-oxoDHA binds to PPARγ and PPARα covalently, making 17-oxoDHA the first of a novel class of PPAR agonists, the PPARα/γ dual covalent agonist. Furthermore, the covalent binding sites were identified as Cys285 for PPARγ and Cys275 for PPARα.
- Laboratory of Drug Design and Medicinal Chemistry, Showa Pharmaceutical University , 3-3165 Higashi-Tamagawagakuen, Machida, Tokyo 194-8543, Japan.
Organizational Affiliation: