Structural Mechanism for Light-driven Transport by a New Type of Chloride Ion Pump, Nonlabens marinus Rhodopsin-3
Hosaka, T., Yoshizawa, S., Nakajima, Y., Ohsawa, N., Hato, M., DeLong, E.F., Kogure, K., Yokoyama, S., Kimura-Someya, T., Iwasaki, W., Shirouzu, M.(2016) J Biol Chem 291: 17488-17495
- PubMed: 27365396
- DOI: https://doi.org/10.1074/jbc.M116.728220
- Primary Citation of Related Structures:
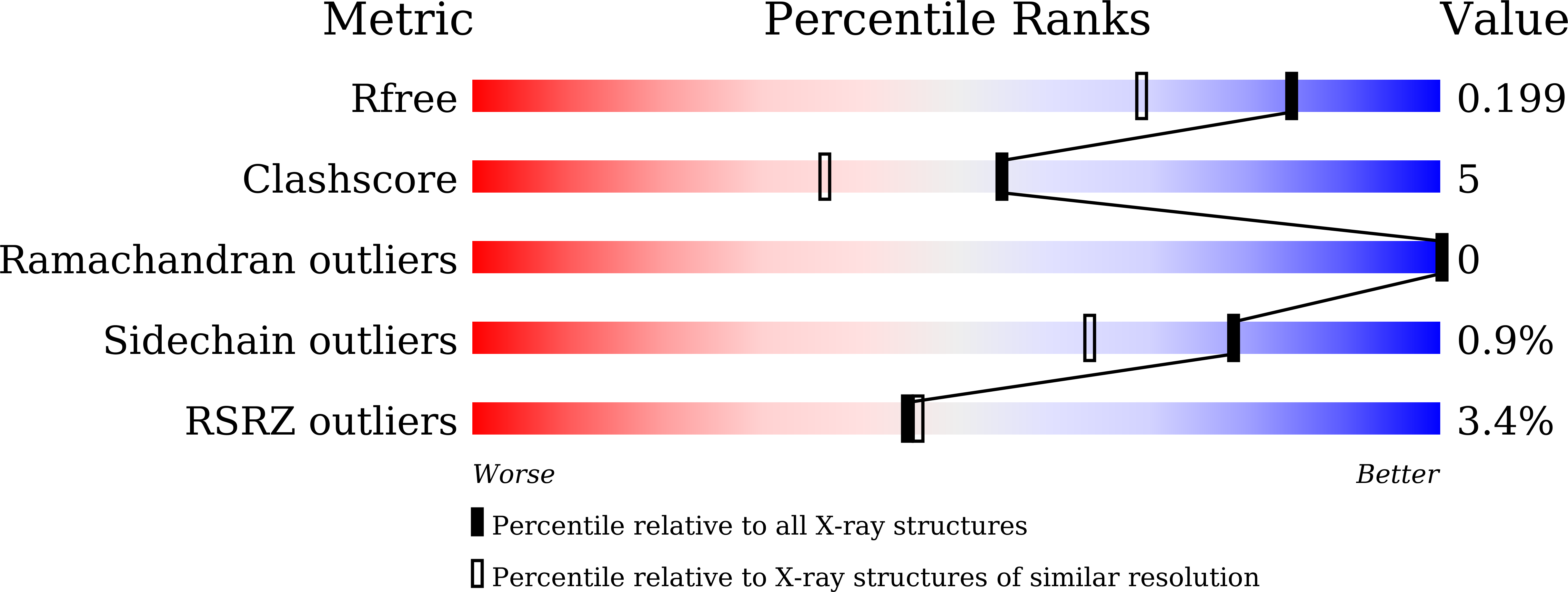
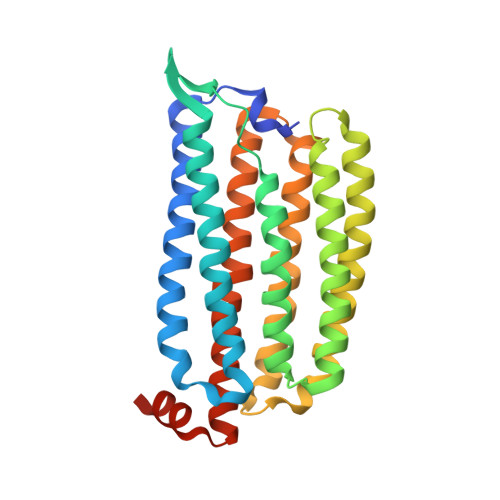
5B2N - PubMed Abstract:
The light-driven inward chloride ion-pumping rhodopsin Nonlabens marinus rhodopsin-3 (NM-R3), from a marine flavobacterium, belongs to a phylogenetic lineage distinct from the halorhodopsins known as archaeal inward chloride ion-pumping rhodopsins. NM-R3 and halorhodopsin have distinct motif sequences that are important for chloride ion binding and transport. In this study, we present the crystal structure of a new type of light-driven chloride ion pump, NM-R3, at 1.58 Å resolution. The structure revealed the chloride ion translocation pathway and showed that a single chloride ion resides near the Schiff base. The overall structure, chloride ion-binding site, and translocation pathway of NM-R3 are different from those of halorhodopsin. Unexpectedly, this NM-R3 structure is similar to the crystal structure of the light-driven outward sodium ion pump, Krokinobacter eikastus rhodopsin 2. Structural and mutational analyses of NM-R3 revealed that most of the important amino acid residues for chloride ion pumping exist in the ion influx region, located on the extracellular side of NM-R3. In contrast, on the opposite side, the cytoplasmic regions of K. eikastus rhodopsin 2 were reportedly important for sodium ion pumping. These results provide new insight into ion selection mechanisms in ion pumping rhodopsins, in which the ion influx regions of both the inward and outward pumps are important for their ion selectivities.
Organizational Affiliation:
From the Division of Structural and Synthetic Biology, RIKEN Center for Life Science Technologies, and.