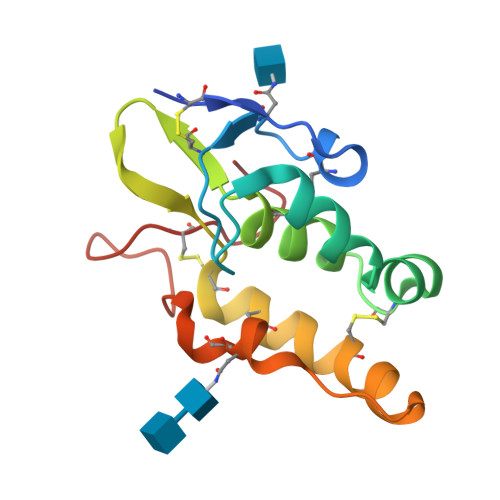
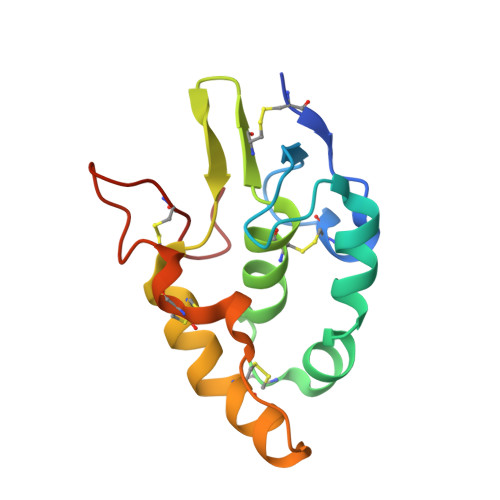
Structural basis of the Norrin-Frizzled 4 interaction.
Shen, G., Ke, J., Wang, Z., Cheng, Z., Gu, X., Wei, Y., Melcher, K., Xu, H.E., Xu, W.(2015) Cell Res 25: 1078-1081
- PubMed: 26227961
- DOI: https://doi.org/10.1038/cr.2015.92
- Primary Citation of Related Structures:
5CL1, 5CM4
Organizational Affiliation:
Department of Biological Structure, University of Washington, Seattle, WA 98195, USA.