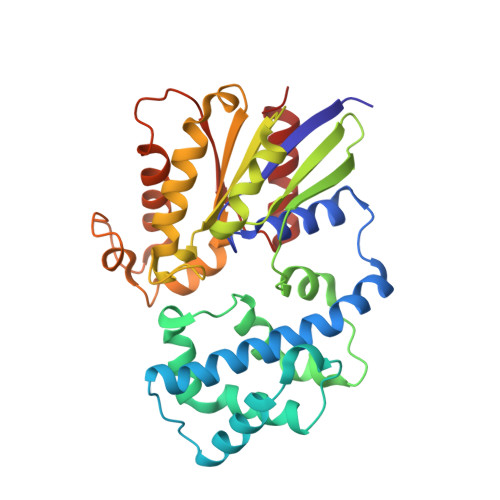
Structure of the Regulator of G Protein Signaling 8 (RGS8)-G alpha q Complex: MOLECULAR BASIS FOR G alpha SELECTIVITY.
Taylor, V.G., Bommarito, P.A., Tesmer, J.J.(2016) J Biological Chem 291: 5138-5145
- PubMed: 26755720
- DOI: https://doi.org/10.1074/jbc.M115.712075
- Primary Citation of Related Structures:
5DO9 - PubMed Abstract:
Regulator of G protein signaling (RGS) proteins interact with activated Gα subunits via their RGS domains and accelerate the hydrolysis of GTP. Although the R4 subfamily of RGS proteins generally accepts both Gαi/o and Gαq/11 subunits as substrates, the R7 and R12 subfamilies select against Gαq/11. In contrast, only one RGS protein, RGS2, is known to be selective for Gαq/11. The molecular basis for this selectivity is not clear. Previously, the crystal structure of RGS2 in complex with Gαq revealed a non-canonical interaction that could be due to interfacial differences imposed by RGS2, the Gα subunit, or both. To resolve this ambiguity, the 2.6 Å crystal structure of RGS8, an R4 subfamily member, was determined in complex with Gαq. RGS8 adopts the same pose on Gαq as it does when bound to Gαi3, indicating that the non-canonical interaction of RGS2 with Gαq is due to unique features of RGS2. Based on the RGS8-Gαq structure, residues in RGS8 that contact a unique α-helical domain loop of Gαq were converted to those typically found in R12 subfamily members, and the reverse substitutions were introduced into RGS10, an R12 subfamily member. Although these substitutions perturbed their ability to stimulate GTP hydrolysis, they did not reverse selectivity. Instead, selectivity for Gαq seems more likely determined by whether strong contacts can be maintained between α6 of the RGS domain and Switch III of Gαq, regions of high sequence and conformational diversity in both protein families.
- From the Life Sciences Institute and the Departments of Pharmacology and Biological Sciences, and the Program in Biophysics, University of Michigan, Ann Arbor, Michigan 48109.
Organizational Affiliation: