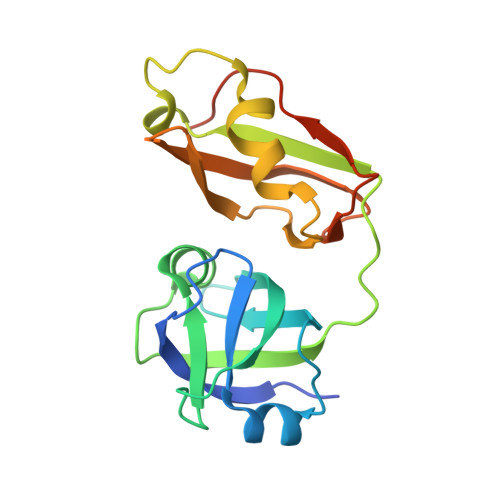
Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Lim, J.J., Lee, Y., Ly, T.T., Kang, J.Y., Lee, J.G., An, J.Y., Youn, H.S., Park, K.R., Kim, T.G., Yang, J.K., Jun, Y., Eom, S.H.(2016) Biochem J 473: 2863-2880
- PubMed: 27407164
- DOI: https://doi.org/10.1042/BCJ20160237
- Primary Citation of Related Structures:
5EPP - PubMed Abstract:
RHBDL4 is an active rhomboid that specifically recognizes and cleaves atypical, positively charged transmembrane endoplasmic reticulum-associated degradation (ERAD) substrates. Interaction of valosin-containing protein (p97/VCP) and RHBDL4 is crucial to retrotranslocate polyubiquitinated substrates for ERAD pathway. Here, we report the first complex structure of VCP-binding motif (VBM) with p97 N-terminal domain (p97N) at 1.88 Å resolution. Consistent with p97 adaptor proteins including p47-ubiquitin regulatory X (UBX), gp78-VCP-interacting motif (VIM), OTU1-UBX-like element, and FAF1-UBX, RHBDL4 VBM also binds at the interface between the two lobes of p97N. Notably, the RF residues in VBM are involved in the interaction with p97N, showing a similar interaction pattern with that of FPR signature motif in the UBX domain, although the directionality is opposite. Comparison of VBM interaction with VIM of gp78, another α-helical motif that interacts with p97N, revealed that the helix direction is inversed. Nevertheless, the conserved arginine residues in both motifs participate in the majority of the interface via extensive hydrogen bonds and ionic interactions with p97N. We identified novel VBM-binding mode to p97N that involves a combination of two types of p97-cofactor specificities observed in the UBX and VIM interactions. This highlights the induced fit model of p97N interdomain cleft upon cofactor binding to form stable p97-cofactor complexes. Our mutational and biochemical analyses in defining the specific interaction between VBM and p97N have elucidated the importance of the highly conserved VBM, applicable to other VBM-containing proteins. We also showed that RHBDL4, ubiquitins, and p97 co-operate for efficient substrate dislocation.
- School of Life Science, Gwangju Institute of Science and Technology (GIST), 123 Cheomdangwagi-ro, Buk-gu, Gwangju 61005, Republic of Korea Steitz Center for Structural Biology, Gwangju Institute of Science and Technology (GIST), 123 Cheomdangwagi-ro, Buk-gu, Gwangju 61005, Republic of Korea.
Organizational Affiliation: