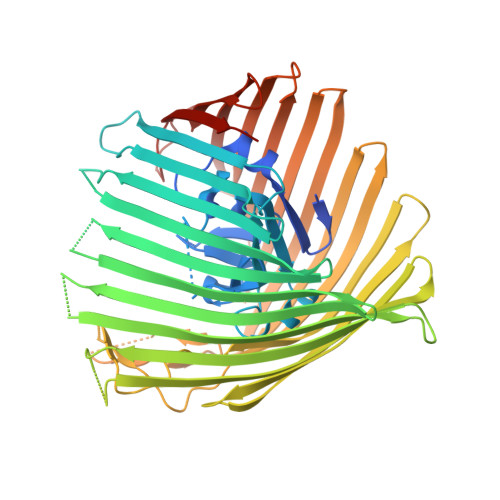
Structure and Function of the PiuA and PirA Siderophore-Drug Receptors from Pseudomonas aeruginosa and Acinetobacter baumannii.
Moynie, L., Luscher, A., Rolo, D., Pletzer, D., Tortajada, A., Weingart, H., Braun, Y., Page, M.G., Naismith, J.H., Kohler, T.(2017) Antimicrob Agents Chemother 61
- PubMed: 28137795
- DOI: https://doi.org/10.1128/AAC.02531-16
- Primary Citation of Related Structures:
5FOK, 5FP1, 5FP2, 5FR8 - PubMed Abstract:
The outer membrane of Gram-negative bacteria presents an efficient barrier to the permeation of antimicrobial molecules. One strategy pursued to circumvent this obstacle is to hijack transport systems for essential nutrients, such as iron. BAL30072 and MC-1 are two monobactams conjugated to a dihydroxypyridone siderophore that are active against Pseudomonas aeruginosa and Acinetobacter baumannii Here, we investigated the mechanism of action of these molecules in A. baumannii We identified two novel TonB-dependent receptors, termed Ab -PiuA and Ab -PirA, that are required for the antimicrobial activity of both agents. Deletion of either piuA or pirA in A. baumannii resulted in 4- to 8-fold-decreased susceptibility, while their overexpression in the heterologous host P. aeruginosa increased susceptibility to the two siderophore-drug conjugates by 4- to 32-fold. The crystal structures of PiuA and PirA from A. baumannii and their orthologues from P. aeruginosa were determined. The structures revealed similar architectures; however, structural differences between PirA and PiuA point to potential differences between their cognate siderophore ligands. Spontaneous mutants, selected upon exposure to BAL30072, harbored frameshift mutations in either the ExbD3 or the TonB3 protein of A. baumannii , forming the cytoplasmic-membrane complex providing the energy for the siderophore translocation process. The results of this study provide insight for the rational design of novel siderophore-drug conjugates against problematic Gram-negative pathogens.
- Biomedical Sciences Research Complex, University of St Andrews, St Andrews, United Kingdom.
Organizational Affiliation: