Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Zebisch, M., Jackson, V.A., Zhao, Y., Jones, E.Y.(2016) Structure 24: 1599
- PubMed: 27524201
- DOI: https://doi.org/10.1016/j.str.2016.06.020
- Primary Citation of Related Structures:
5FWS, 5FWT, 5FWU, 5FWV, 5FWW - PubMed Abstract:
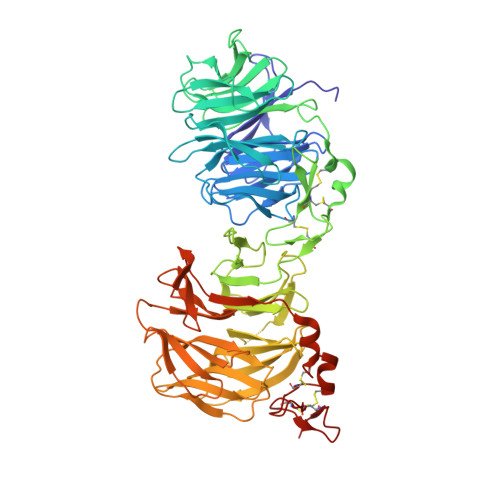
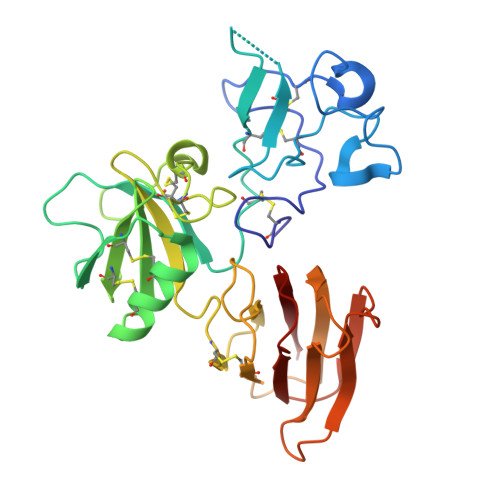
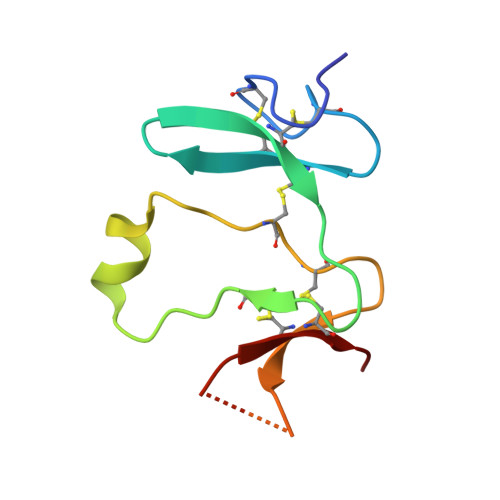
Kremen 1 and 2 have been identified as co-receptors for Dickkopf (Dkk) proteins, hallmark secreted antagonists of canonical Wnt signaling. We present here three crystal structures of the ectodomain of human Kremen1 (KRM1ECD) at resolutions between 1.9 and 3.2 Å. KRM1ECD emerges as a rigid molecule with tight interactions stabilizing a triangular arrangement of its Kringle, WSC, and CUB structural domains. The structures reveal an unpredicted homology of the WSC domain to hepatocyte growth factor. We further report the general architecture of the ternary complex formed by the Wnt co-receptor Lrp5/6, Dkk, and Krm, determined from a low-resolution complex crystal structure between β-propeller/EGF repeats (PE) 3 and 4 of the Wnt co-receptor LRP6 (LRP6PE3PE4), the cysteine-rich domain 2 (CRD2) of DKK1, and KRM1ECD. DKK1CRD2 is sandwiched between LRP6PE3 and KRM1Kringle-WSC. Modeling studies supported by surface plasmon resonance suggest a direct interaction site between Krm1CUB and Lrp6PE2.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK. Electronic address: matthias.zebisch@evotec.com.
Organizational Affiliation: