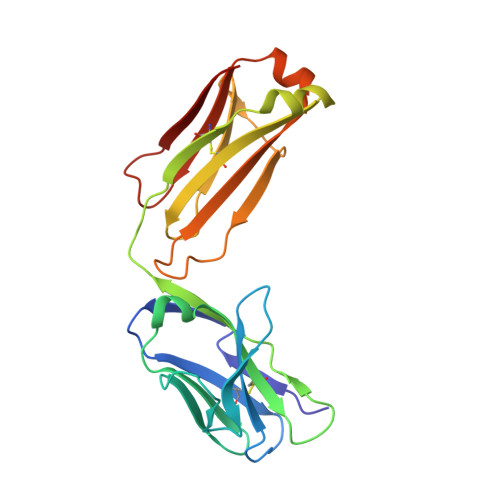
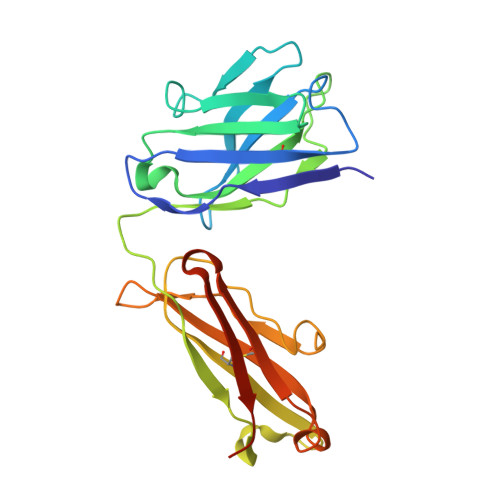
Structural diversity in a human antibody germline library.
Teplyakov, A., Obmolova, G., Malia, T.J., Luo, J., Muzammil, S., Sweet, R., Almagro, J.C., Gilliland, G.L.(2016) MAbs 8: 1045-1063
- PubMed: 27210805
- DOI: https://doi.org/10.1080/19420862.2016.1190060
- Primary Citation of Related Structures:
5I15, 5I16, 5I17, 5I18, 5I19, 5I1A, 5I1C, 5I1D, 5I1E, 5I1G, 5I1H, 5I1I, 5I1J, 5I1K, 5I1L - PubMed Abstract:
To support antibody therapeutic development, the crystal structures of a set of 16 germline variants composed of 4 different kappa light chains paired with 4 different heavy chains have been determined. All four heavy chains of the antigen-binding fragments (Fabs) have the same complementarity-determining region (CDR) H3 that was reported in an earlier Fab structure. The structure analyses include comparisons of the overall structures, canonical structures of the CDRs and the VH:VL packing interactions. The CDR conformations for the most part are tightly clustered, especially for the ones with shorter lengths. The longer CDRs with tandem glycines or serines have more conformational diversity than the others. CDR H3, despite having the same amino acid sequence, exhibits the largest conformational diversity. About half of the structures have CDR H3 conformations similar to that of the parent; the others diverge significantly. One conclusion is that the CDR H3 conformations are influenced by both their amino acid sequence and their structural environment determined by the heavy and light chain pairing. The stem regions of 14 of the variant pairs are in the 'kinked' conformation, and only 2 are in the extended conformation. The packing of the VH and VL domains is consistent with our knowledge of antibody structure, and the tilt angles between these domains cover a range of 11 degrees. Two of 16 structures showed particularly large variations in the tilt angles when compared with the other pairings. The structures and their analyses provide a rich foundation for future antibody modeling and engineering efforts.
- a Janssen Research & Development LLC, Spring House , PA , USA.
Organizational Affiliation: