Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and PDZRhoGEF
Wang, R., Yan, Z., Lv, Z., Ma, L., Miao, Q., Chen, Q., Wang, F., Li, J., Miao, L.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Rho guanine nucleotide exchange factor 11 | A, C [auth E] | 369 | Homo sapiens | Mutation(s): 0 Gene Names: ARHGEF11, KIAA0380 | 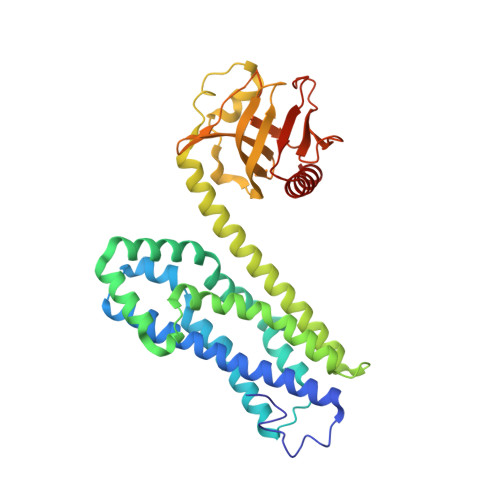 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O15085 (Homo sapiens) Explore O15085 Go to UniProtKB: O15085 | |||||
PHAROS: O15085 GTEx: ENSG00000132694 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O15085 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transforming protein RhoA | B, D [auth F] | 181 | Homo sapiens | Mutation(s): 0 Gene Names: RHOA, ARH12, ARHA, RHO12 EC: 3.6.5.2 | 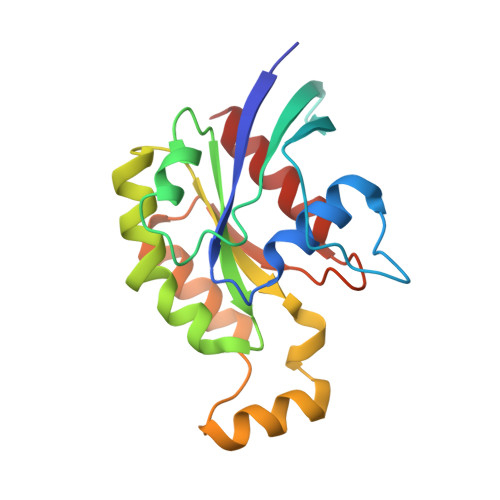 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61586 (Homo sapiens) Explore P61586 Go to UniProtKB: P61586 | |||||
PHAROS: P61586 GTEx: ENSG00000067560 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61586 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RA0 Query on RA0 | G [auth B], I [auth F] | 3-{3-[ethyl(quinolin-2-yl)amino]phenyl}propanoic acid C20 H20 N2 O2 CGGUWDMVJGUARS-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | E [auth A], F [auth A], H [auth E] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 88.855 | α = 90 |
| b = 119.049 | β = 114.39 |
| c = 90.526 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Jiangsu Province science and technology plan items (Jiangsu Province science and technology enterprise technology innovation Fund) | China | BC2013062 |