Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Lu, J., Chan, C., Yu, L., Fan, J., Sun, F., Zhai, Y.(2020) Commun Biol
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein UPS1, mitochondrial | 189 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: UPS1, YLR193C | 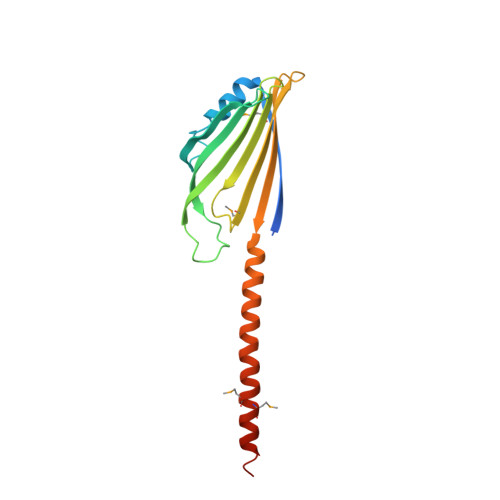 | |
UniProt | |||||
Find proteins for Q05776 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q05776 Go to UniProtKB: Q05776 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q05776 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial distribution and morphology protein 35 | 86 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: MDM35, YKL053C-A | 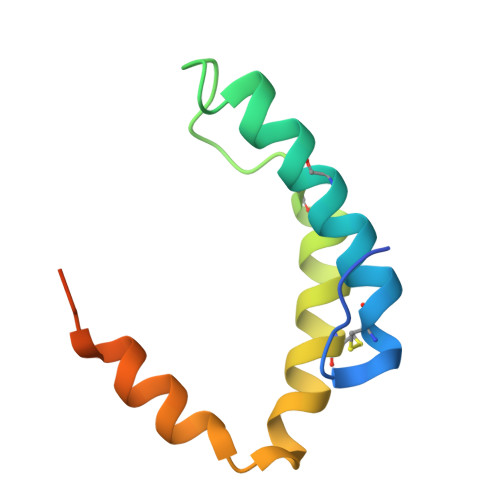 | |
UniProt | |||||
Find proteins for O60200 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore O60200 Go to UniProtKB: O60200 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60200 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein UPS1, mitochondrial | 189 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: UPS1, YLR193C |  | |
UniProt | |||||
Find proteins for Q05776 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q05776 Go to UniProtKB: Q05776 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q05776 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein UPS1, mitochondrial | 189 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: UPS1, YLR193C | 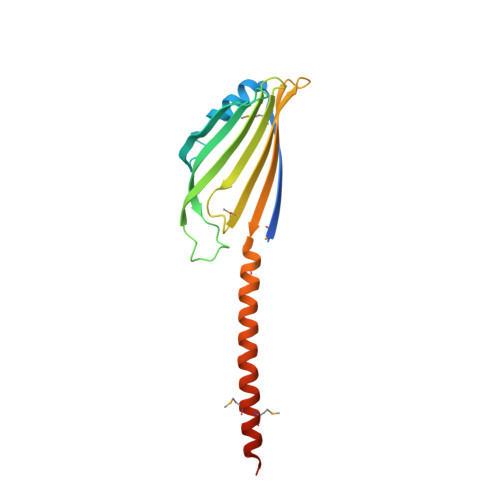 | |
UniProt | |||||
Find proteins for Q05776 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q05776 Go to UniProtKB: Q05776 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q05776 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial distribution and morphology protein 35 | 86 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: MDM35, YKL053C-A | 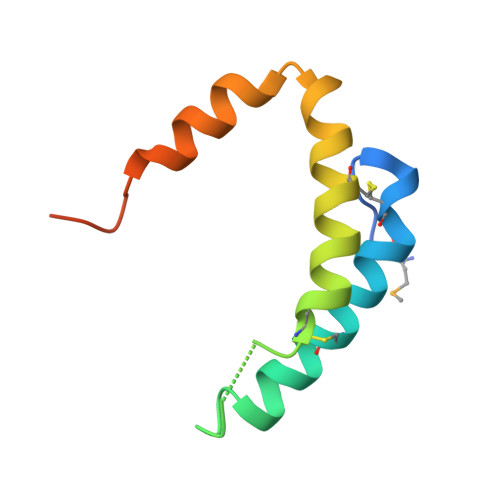 | |
UniProt | |||||
Find proteins for O60200 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore O60200 Go to UniProtKB: O60200 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60200 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, E, I, K | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 83.311 | α = 90 |
| b = 130.736 | β = 103.09 |
| c = 84.271 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| China | -- |