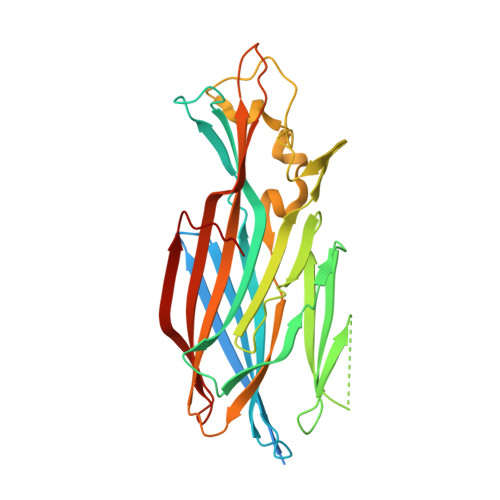
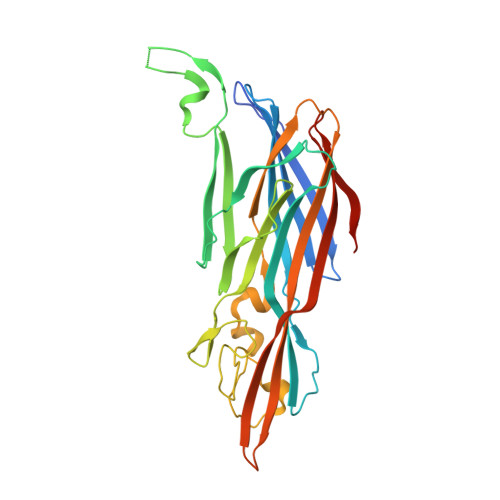
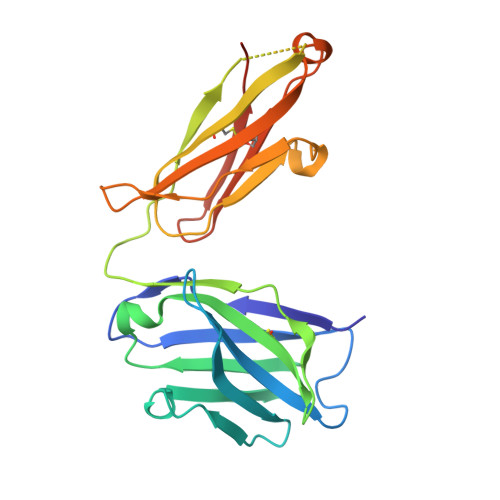
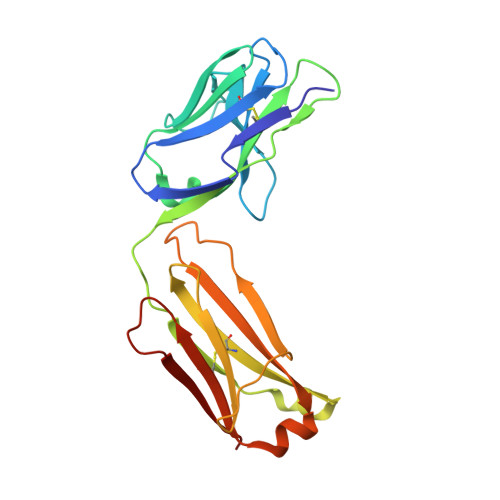
Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Badarau, A., Rouha, H., Malafa, S., Battles, M.B., Walker, L., Nielson, N., Dolezilkova, I., Teubenbacher, A., Banerjee, S., Maierhofer, B., Weber, S., Stulik, L., Logan, D.T., Welin, M., Mirkina, I., Pleban, C., Zauner, G., Gross, K., Jagerhofer, M., Magyarics, Z., Nagy, E.(2016) MAbs 8: 1347-1360
- PubMed: 27467113
- DOI: https://doi.org/10.1080/19420862.2016.1215791
- Primary Citation of Related Structures:
5K59 - PubMed Abstract:
LukGH (LukAB) is a potent leukocidin of Staphylococcus aureus that lyses human phagocytic cells and is thought to contribute to immune evasion. Unlike the other bi-component leukocidins of S. aureus, LukGH forms a heterodimer before binding to its receptor, CD11b expressed on professional phagocytic cells, and displays significant sequence variation. We employed a high diversity human IgG1 library presented on yeast cells to discover monoclonal antibodies (mAbs) neutralizing the cytolytic activity of LukGH. Recombinant LukG and LukH monomers or a LukGH dimer were used as capture antigens in the library selections. We found that mAbs identified with LukG or LukH as bait had no or very low toxin neutralization potency. In contrast, LukGH dimer-selected antibodies proved to be highly potent, and several mAbs were able to neutralize even the most divergent LukGH variants. Based on biolayer interferometry and mesoscale discovery, the high affinity antibody binding site on the LukGH complex was absent on the individual monomers, suggesting that it was generated upon formation of the LukG-LukH dimer. X-ray crystallography analysis of the complex between the LukGH dimer and the antigen-binding fragment of a very potent mAb (PDB code 5K59) indicated that the epitope is located in the predicted cell binding region (rim domain) of LukGH. The corresponding IgG inhibited the binding of LukGH dimer to target cells. Our data suggest that knowledge of the native conformation of target molecules is essential to generate high affinity and functional mAbs.
- a Arsanis Biosciences , Campus Vienna Biocenter, Vienna , Austria.
Organizational Affiliation: