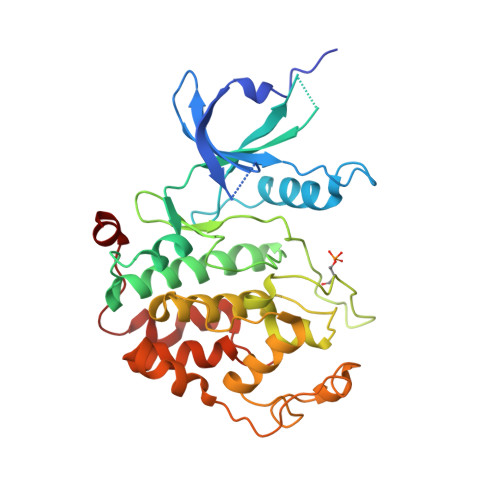
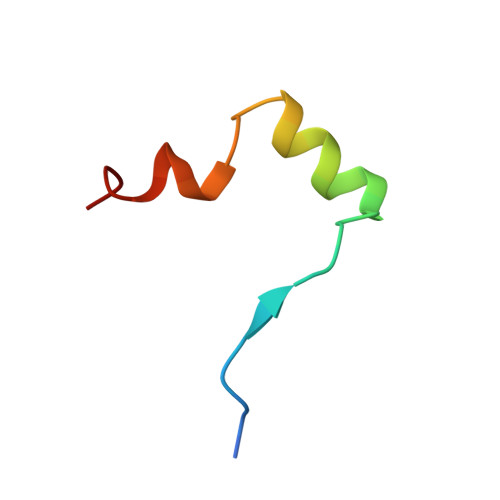
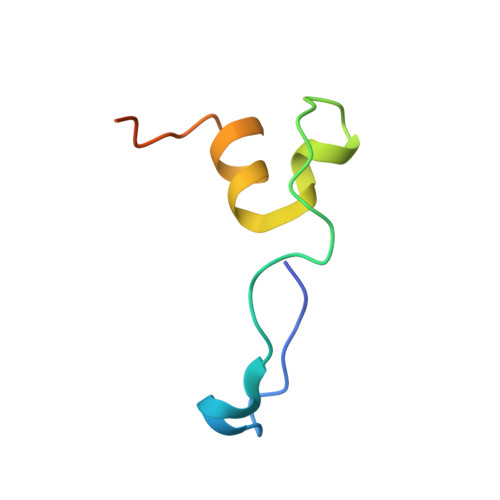
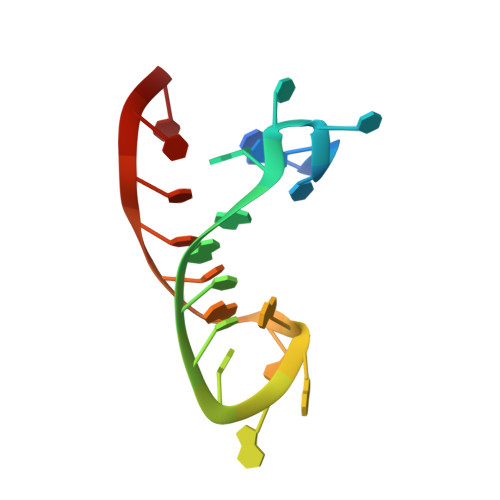
Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Schulze-Gahmen, U., Echeverria, I., Stjepanovic, G., Bai, Y., Lu, H., Schneidman-Duhovny, D., Doudna, J.A., Zhou, Q., Sali, A., Hurley, J.H.(2016) Elife 5
- PubMed: 27731797
- DOI: https://doi.org/10.7554/eLife.15910
- Primary Citation of Related Structures:
5L1Z - PubMed Abstract:
HIV-1 Tat hijacks the human superelongation complex (SEC) to promote proviral transcription. Here we report the 5.9 Å structure of HIV-1 TAR in complex with HIV-1 Tat and human AFF4, CDK9, and CycT1. The TAR central loop contacts the CycT1 Tat-TAR recognition motif (TRM) and the second Tat Zn 2+ -binding loop. Hydrogen-deuterium exchange (HDX) shows that AFF4 helix 2 is stabilized in the TAR complex despite not touching the RNA, explaining how it enhances TAR binding to the SEC 50-fold. RNA SHAPE and SAXS data were used to help model the extended (Tat Arginine-Rich Motif) ARM, which enters the TAR major groove between the bulge and the central loop. The structure and functional assays collectively support an integrative structure and a bipartite binding model, wherein the TAR central loop engages the CycT1 TRM and compact core of Tat, while the TAR major groove interacts with the extended Tat ARM.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States.
Organizational Affiliation: