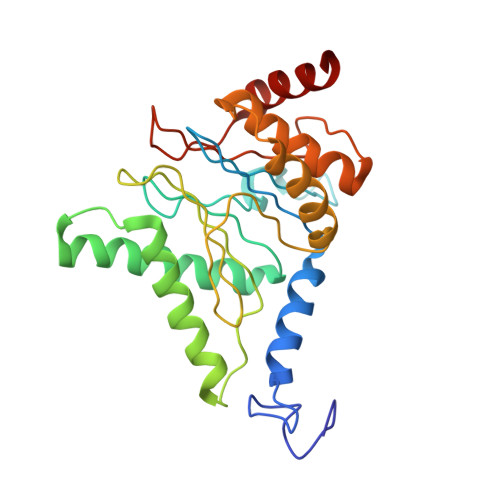
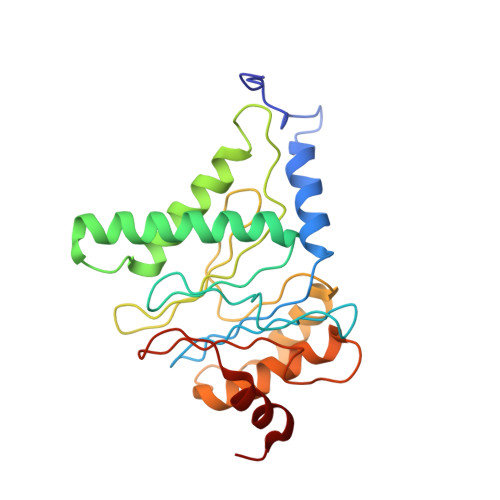
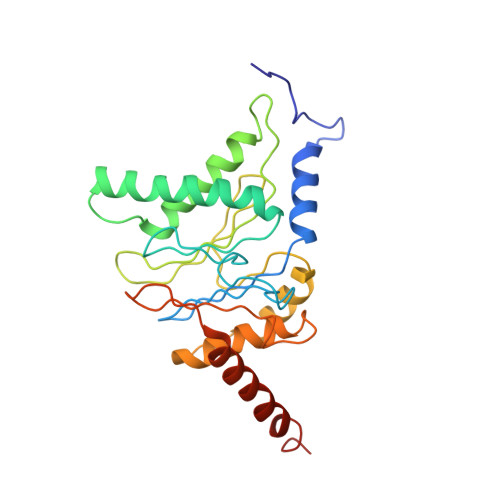
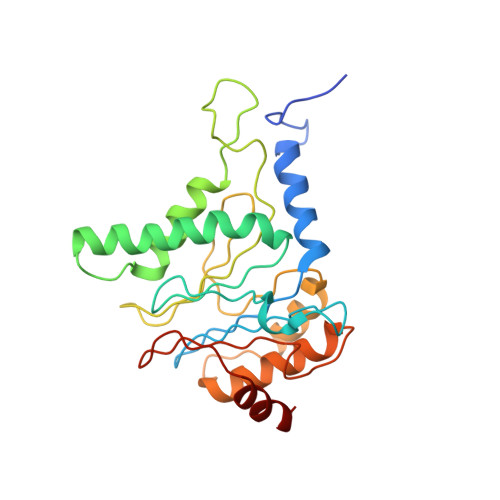
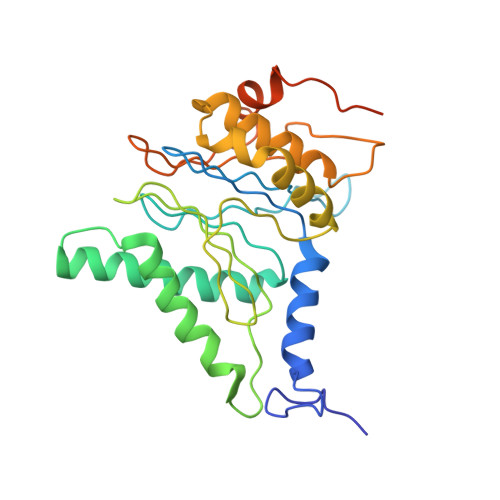
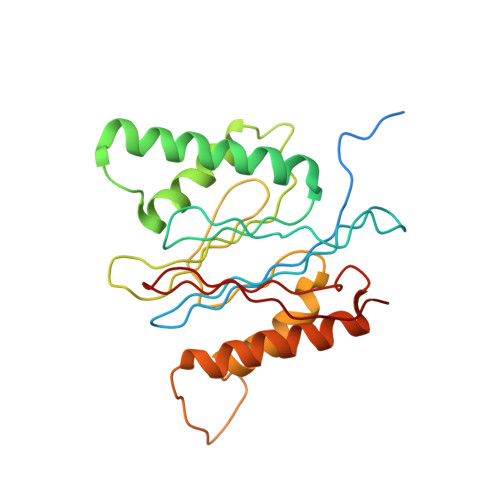
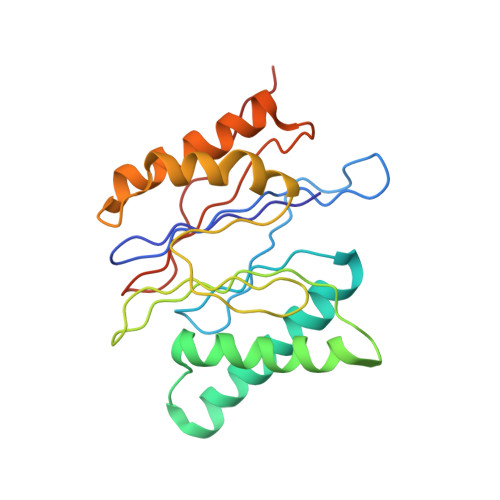
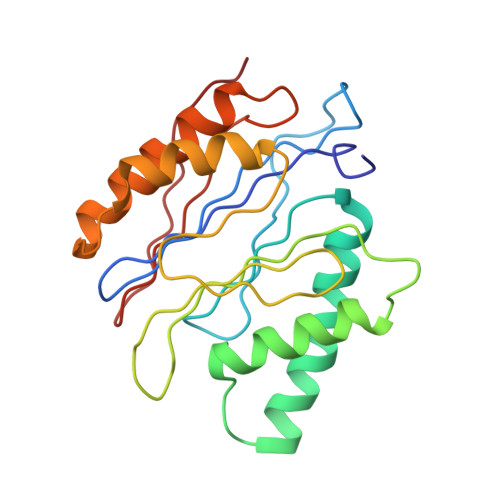
Structure of the human 26S proteasome at a resolution of 3.9 angstrom.
Schweitzer, A., Aufderheide, A., Rudack, T., Beck, F., Pfeifer, G., Plitzko, J.M., Sakata, E., Schulten, K., Forster, F., Baumeister, W.(2016) Proc Natl Acad Sci U S A 113: 7816-7821
- PubMed: 27342858
- DOI: https://doi.org/10.1073/pnas.1608050113
- Primary Citation of Related Structures:
5L4G, 5L4K - PubMed Abstract:
Protein degradation in eukaryotic cells is performed by the Ubiquitin-Proteasome System (UPS). The 26S proteasome holocomplex consists of a core particle (CP) that proteolytically degrades polyubiquitylated proteins, and a regulatory particle (RP) containing the AAA-ATPase module. This module controls access to the proteolytic chamber inside the CP and is surrounded by non-ATPase subunits (Rpns) that recognize substrates and deubiquitylate them before unfolding and degradation. The architecture of the 26S holocomplex is highly conserved between yeast and humans. The structure of the human 26S holocomplex described here reveals previously unidentified features of the AAA-ATPase heterohexamer. One subunit, Rpt6, has ADP bound, whereas the other five have ATP in their binding pockets. Rpt6 is structurally distinct from the other five Rpt subunits, most notably in its pore loop region. For Rpns, the map reveals two main, previously undetected, features: the C terminus of Rpn3 protrudes into the mouth of the ATPase ring; and Rpn1 and Rpn2, the largest proteasome subunits, are linked by an extended connection. The structural features of the 26S proteasome observed in this study are likely to be important for coordinating the proteasomal subunits during substrate processing.
- Department of Molecular Structural Biology, Max-Planck Institute of Biochemistry, D-82152 Martinsried, Germany;
Organizational Affiliation: