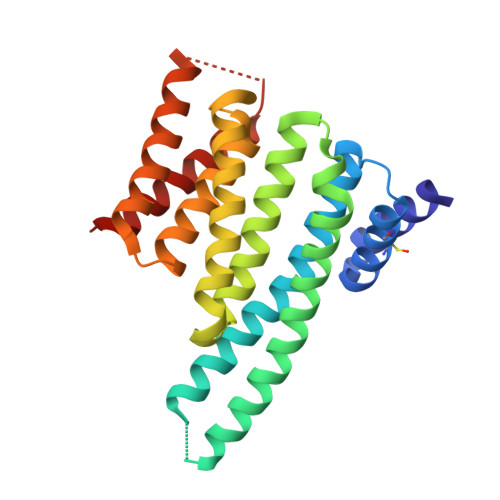
The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
Bier, D., Mittal, S., Bravo-Rodriguez, K., Sowislok, A., Guillory, X., Briels, J., Heid, C., Bartel, M., Wettig, B., Brunsveld, L., Sanchez-Garcia, E., Schrader, T., Ottmann, C.(2017) J Am Chem Soc 139: 16256-16263
- PubMed: 29039919
- DOI: https://doi.org/10.1021/jacs.7b07939
- Primary Citation of Related Structures:
5M35, 5M36, 5M37 - PubMed Abstract:
Protein regions that are involved in protein-protein interactions (PPIs) very often display a high degree of intrinsic disorder, which is reduced during the recognition process. A prime example is binding of the rigid 14-3-3 adapter proteins to their numerous partner proteins, whose recognition motifs undergo an extensive disorder-to-order transition. In this context, it is highly desirable to control this entropy-costly process using tailored stabilizing agents. This study reveals how the molecular tweezer CLR01 tunes the 14-3-3/Cdc25CpS216 protein-protein interaction. Protein crystallography, biophysical affinity determination and biomolecular simulations unanimously deliver a remarkable finding: a supramolecular "Janus" ligand can bind simultaneously to a flexible peptidic PPI recognition motif and to a well-structured adapter protein. This binding fills a gap in the protein-protein interface, "freezes" one of the conformational states of the intrinsically disordered Cdc25C protein partner and enhances the apparent affinity of the interaction. This is the first structural and functional proof of a supramolecular ligand targeting a PPI interface and stabilizing the binding of an intrinsically disordered recognition motif to a rigid partner protein.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology , Den Dolech 2, 5612 AZ Eindhoven, The Netherlands.
Organizational Affiliation: