Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Ge, C., Xu, B., Liang, B., Lonnblom, E., Lundstrom, S.L., Zubarev, R.A., Ayoglu, B., Nilsson, P., Skogh, T., Kastbom, A., Malmstrom, V., Klareskog, L., Toes, R.E.M., Rispens, T., Dobritzsch, D., Holmdahl, R.(2019) Arthritis Rheumatol 71: 210-221
- PubMed: 30152126
- DOI: https://doi.org/10.1002/art.40698
- Primary Citation of Related Structures:
5OCK, 5OCX, 5OCY, 5OD0, 5OD8, 5ODB - PubMed Abstract:
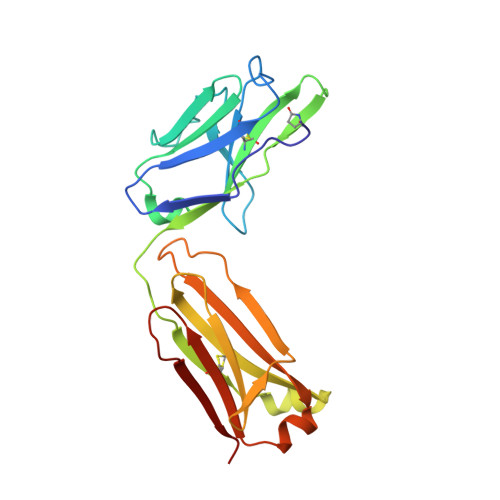
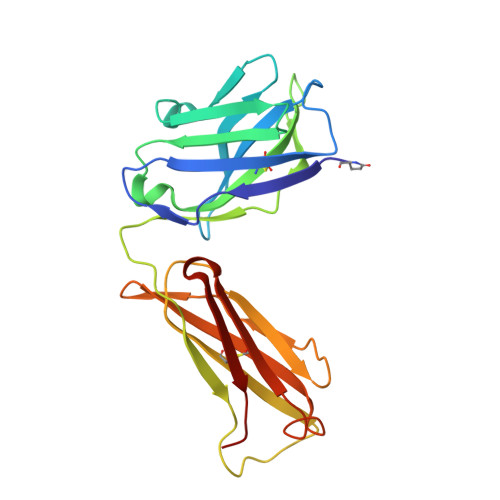
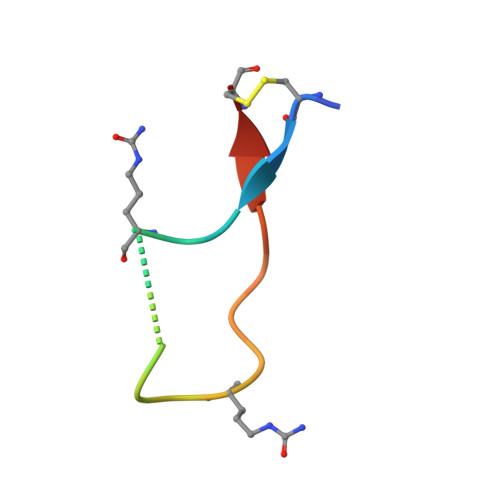
Anti-citrullinated protein antibodies (ACPAs) develop many years before the clinical onset of rheumatoid arthritis (RA). This study was undertaken to address the molecular basis of the specificity and cross-reactivity of ACPAs from patients with RA. Antibodies isolated from RA patients were expressed as monoclonal chimeric antibodies with mouse Fc. These antibodies were characterized for glycosylation using mass spectrometry, and their cross-reactivity was assessed using Biacore and Luminex immunoassays. The crystal structures of the antigen-binding fragment (Fab) of the monoclonal ACPA E4 in complex with 3 different citrullinated peptides were determined using x-ray crystallography. The prevalence of autoantibodies reactive against 3 of the citrullinated peptides that also interacted with E4 was investigated by Luminex immunoassay in 2 Swedish cohorts of RA patients. Analysis of the crystal structures of a monoclonal ACPA from human RA serum in complex with citrullinated peptides revealed key residues of several complementarity-determining regions that recognized the citrulline as well as the neighboring peptide backbone, but with limited contact with the side chains of the peptides. The same citrullinated peptides were recognized by high titers of serum autoantibodies in 2 large cohorts of RA patients. These data show, for the first time, how ACPAs derived from human RA serum recognize citrulline. The specific citrulline recognition and backbone-mediated interactions provide a structural explanation for the promiscuous recognition of citrullinated peptides by RA-specific ACPAs.
- Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: