Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene.
Lilley, C.J., Maqbool, A., Wu, D., Yusup, H.B., Jones, L.M., Birch, P.R.J., Banfield, M.J., Urwin, P.E., Eves-van den Akker, S.(2018) PLoS Genet 14: e1007310-e1007310
- PubMed: 29641602
- DOI: https://doi.org/10.1371/journal.pgen.1007310
- Primary Citation of Related Structures:
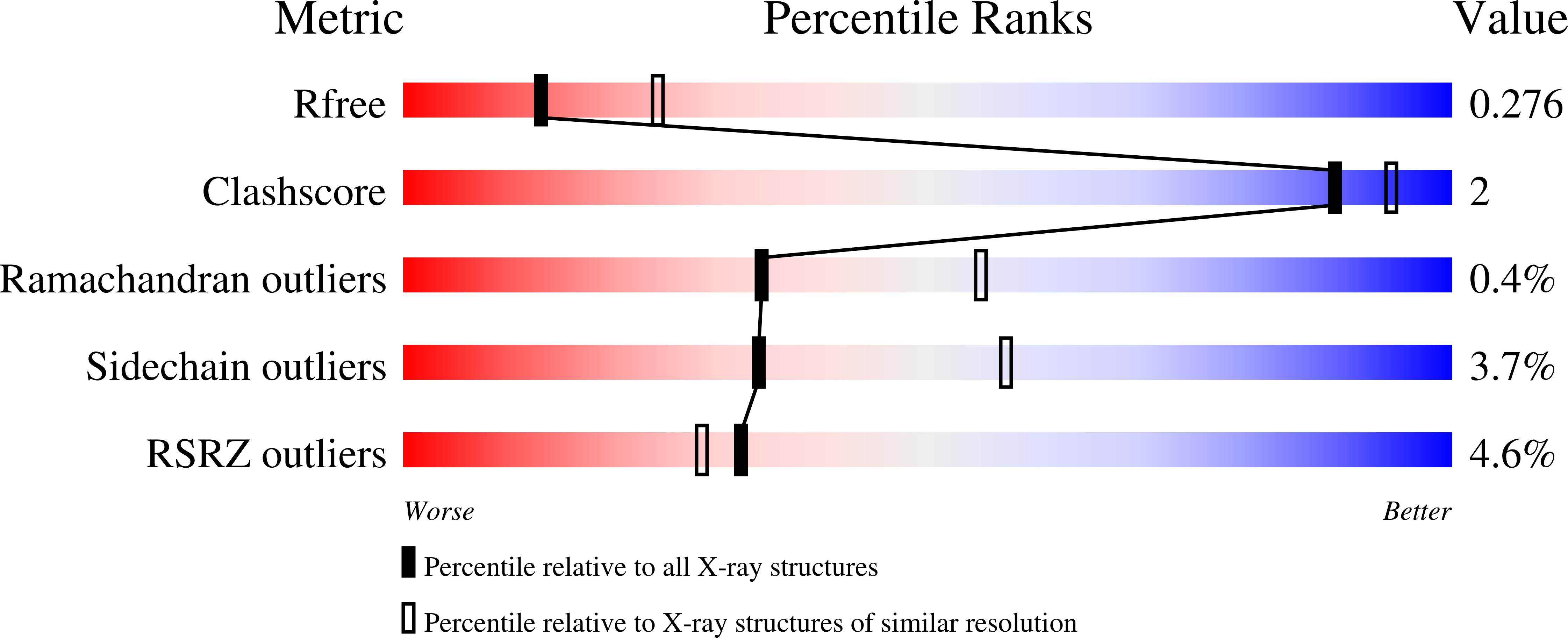
5OES, 5OET, 5OEU, 5OEV - PubMed Abstract:
Plant pathogens and parasites are a major threat to global food security. Plant parasitism has arisen four times independently within the phylum Nematoda, resulting in at least one parasite of every major food crop in the world. Some species within the most economically important order (Tylenchida) secrete proteins termed effectors into their host during infection to re-programme host development and immunity. The precise detail of how nematodes evolve new effectors is not clear. Here we reconstruct the evolutionary history of a novel effector gene family. We show that during the evolution of plant parasitism in the Tylenchida, the housekeeping glutathione synthetase (GS) gene was extensively replicated. New GS paralogues acquired multiple dorsal gland promoter elements, altered spatial expression to the secretory dorsal gland, altered temporal expression to primarily parasitic stages, and gained a signal peptide for secretion. The gene products are delivered into the host plant cell during infection, giving rise to "GS-like effectors". Remarkably, by solving the structure of GS-like effectors we show that during this process they have also diversified in biochemical activity, and likely represent the founding members of a novel class of GS-like enzyme. Our results demonstrate the re-purposing of an endogenous housekeeping gene to form a family of effectors with modified functions. We anticipate that our discovery will be a blueprint to understand the evolution of other plant-parasitic nematode effectors, and the foundation to uncover a novel enzymatic function.
Organizational Affiliation:
Centre for Plant Sciences, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom.