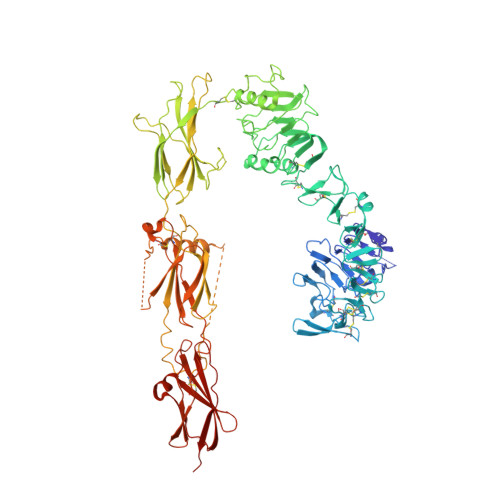
How ligand binds to the type 1 insulin-like growth factor receptor.
Xu, Y., Kong, G.K., Menting, J.G., Margetts, M.B., Delaine, C.A., Jenkin, L.M., Kiselyov, V.V., De Meyts, P., Forbes, B.E., Lawrence, M.C.(2018) Nat Commun 9: 821-821
- PubMed: 29483580
- DOI: https://doi.org/10.1038/s41467-018-03219-7
- Primary Citation of Related Structures:
5U8Q, 5U8R - PubMed Abstract:
Human type 1 insulin-like growth factor receptor is a homodimeric receptor tyrosine kinase that signals into pathways directing normal cellular growth, differentiation and proliferation, with aberrant signalling implicated in cancer. Insulin-like growth factor binding is understood to relax conformational restraints within the homodimer, initiating transphosphorylation of the tyrosine kinase domains. However, no three-dimensional structures exist for the receptor ectodomain to inform atomic-level understanding of these events. Here, we present crystal structures of the ectodomain in apo form and in complex with insulin-like growth factor I, the latter obtained by crystal soaking. These structures not only provide a wealth of detail of the growth factor interaction with the receptor's primary ligand-binding site but also indicate that ligand binding separates receptor domains by a mechanism of induced fit. Our findings are of importance to the design of agents targeting IGF-1R and its partner protein, the human insulin receptor.
- The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC, 3052, Australia.
Organizational Affiliation: