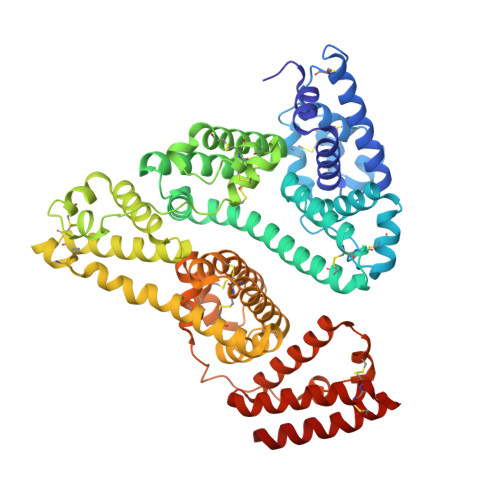
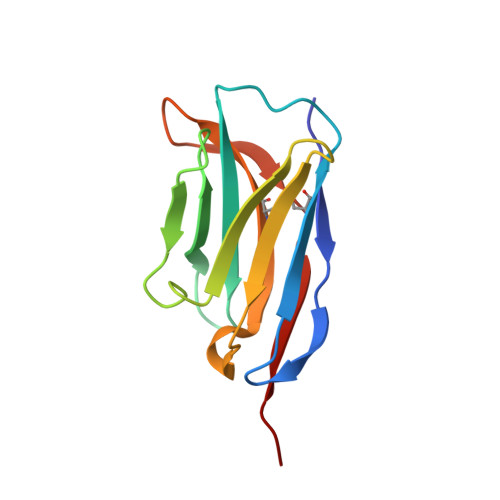
Yeast surface display platform for rapid discovery of conformationally selective nanobodies.
McMahon, C., Baier, A.S., Pascolutti, R., Wegrecki, M., Zheng, S., Ong, J.X., Erlandson, S.C., Hilger, D., Rasmussen, S.G.F., Ring, A.M., Manglik, A., Kruse, A.C.(2018) Nat Struct Mol Biol 25: 289-296
- PubMed: 29434346
- DOI: https://doi.org/10.1038/s41594-018-0028-6
- Primary Citation of Related Structures:
5VNV, 5VNW - PubMed Abstract:
Camelid single-domain antibody fragments ('nanobodies') provide the remarkable specificity of antibodies within a single 15-kDa immunoglobulin V HH domain. This unique feature has enabled applications ranging from use as biochemical tools to therapeutic agents. Nanobodies have emerged as especially useful tools in protein structural biology, facilitating studies of conformationally dynamic proteins such as G-protein-coupled receptors (GPCRs). Nearly all nanobodies available to date have been obtained by animal immunization, a bottleneck restricting many applications of this technology. To solve this problem, we report a fully in vitro platform for nanobody discovery based on yeast surface display. We provide a blueprint for identifying nanobodies, demonstrate the utility of the library by crystallizing a nanobody with its antigen, and most importantly, we utilize the platform to discover conformationally selective nanobodies to two distinct human GPCRs. To facilitate broad deployment of this platform, the library and associated protocols are freely available for nonprofit research.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: