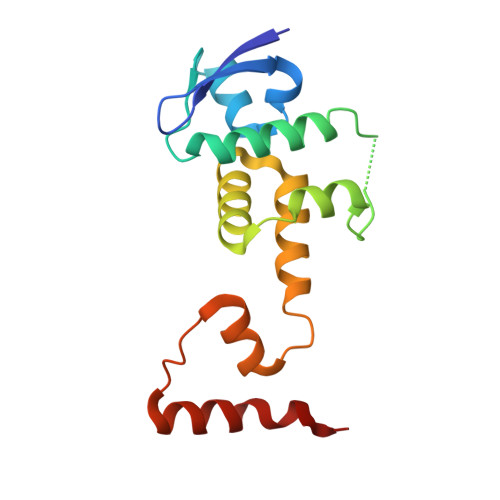
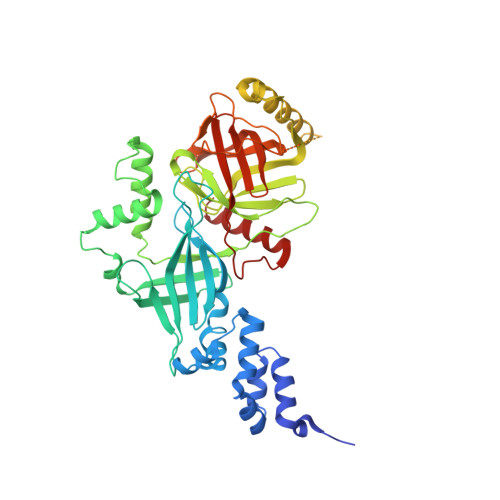
Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Li, Y., Jin, K., Bunker, E., Zhang, X., Luo, X., Liu, X., Hao, B.(2018) Proc Natl Acad Sci U S A 115: 319-324
- PubMed: 29279382
- DOI: https://doi.org/10.1073/pnas.1708677115
- Primary Citation of Related Structures:
5VZT, 5VZU - PubMed Abstract:
Ubiquitin-dependent proteolysis of cyclin D1 is associated with normal and tumor cell proliferation and survival. The SCF FBXO31 (Skp1-Cul1-Rbx1-FBXO31) ubiquitin ligase complex mediates genotoxic stress-induced cyclin D1 degradation. Previous studies have suggested that cyclin D1 levels are maintained at steady state by phosphorylation-dependent nuclear export and subsequent proteolysis in the cytoplasm. Here we present the crystal structures of the Skp1-FBXO31 complex alone and bound to a phosphorylated cyclin D1 C-terminal peptide. FBXO31 possesses a unique substrate-binding domain consisting of two β-barrel motifs, whereas cyclin D1 binds to FBXO31 by tucking its free C-terminal carboxylate tail into an open cavity of the C-terminal FBXO31 β-barrel. Biophysical and functional studies demonstrate that SCF FBXO31 is capable of recruiting and ubiquitinating cyclin D1 in a phosphorylation-independent manner. Our findings provide a conceptual framework for understanding the substrate specificity of the F-box protein FBXO31 and the mechanism of FBXO31-regulated cyclin D1 protein turnover.
- Department of Molecular Biology and Biophysics, University of Connecticut Health Center, Farmington, CT 06030.
Organizational Affiliation: