Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution
Iqbal, N., Kaur, P., Sharma, S., Singh, T.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3-dehydroquinate dehydratase | 148 | Acinetobacter baumannii ATCC 17978 | Mutation(s): 0 Gene Names: aroQ, A1S_2009 EC: 4.2.1.10 | 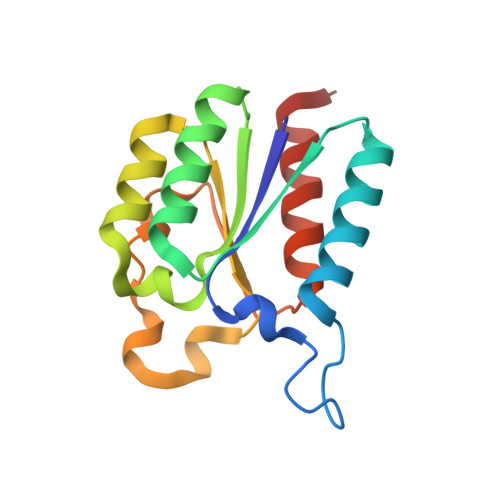 | |
UniProt | |||||
Find proteins for A3M692 (Acinetobacter baumannii (strain ATCC 17978 / DSM 105126 / CIP 53.77 / LMG 1025 / NCDC KC755 / 5377)) Explore A3M692 Go to UniProtKB: A3M692 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A3M692 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DQA Query on DQA | F [auth A], H [auth B], J [auth C], L [auth D] | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID C7 H10 O6 WVMWZWGZRAXUBK-SYTVJDICSA-N |  | ||
| NA Query on NA | E [auth A], G [auth B], I [auth C], K [auth D] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 139.117 | α = 90 |
| b = 139.117 | β = 90 |
| c = 79.587 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |