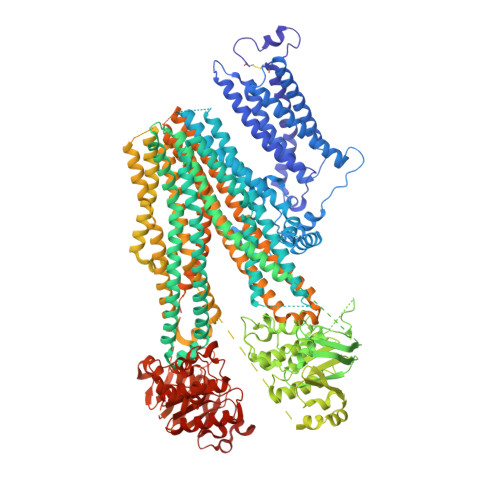
Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Martin, G.M., Kandasamy, B., DiMaio, F., Yoshioka, C., Shyng, S.L.(2017) Elife 6
- PubMed: 29035201
- DOI: https://doi.org/10.7554/eLife.31054
- Primary Citation of Related Structures:
6BAA - PubMed Abstract:
Sulfonylureas are anti-diabetic medications that act by inhibiting pancreatic K ATP channels composed of SUR1 and Kir6.2. The mechanism by which these drugs interact with and inhibit the channel has been extensively investigated, yet it remains unclear where the drug binding pocket resides. Here, we present a cryo-EM structure of a hamster SUR1/rat Kir6.2 channel bound to a high-affinity sulfonylurea drug glibenclamide and ATP at 3.63 Å resolution, which reveals unprecedented details of the ATP and glibenclamide binding sites. Importantly, the structure shows for the first time that glibenclamide is lodged in the transmembrane bundle of the SUR1-ABC core connected to the first nucleotide binding domain near the inner leaflet of the lipid bilayer. Mutation of residues predicted to interact with glibenclamide in our model led to reduced sensitivity to glibenclamide. Our structure provides novel mechanistic insights of how sulfonylureas and ATP interact with the K ATP channel complex to inhibit channel activity.
- Department of Biochemistry and Molecular Biology, Oregon Health and Science University, Portland, United States.
Organizational Affiliation: