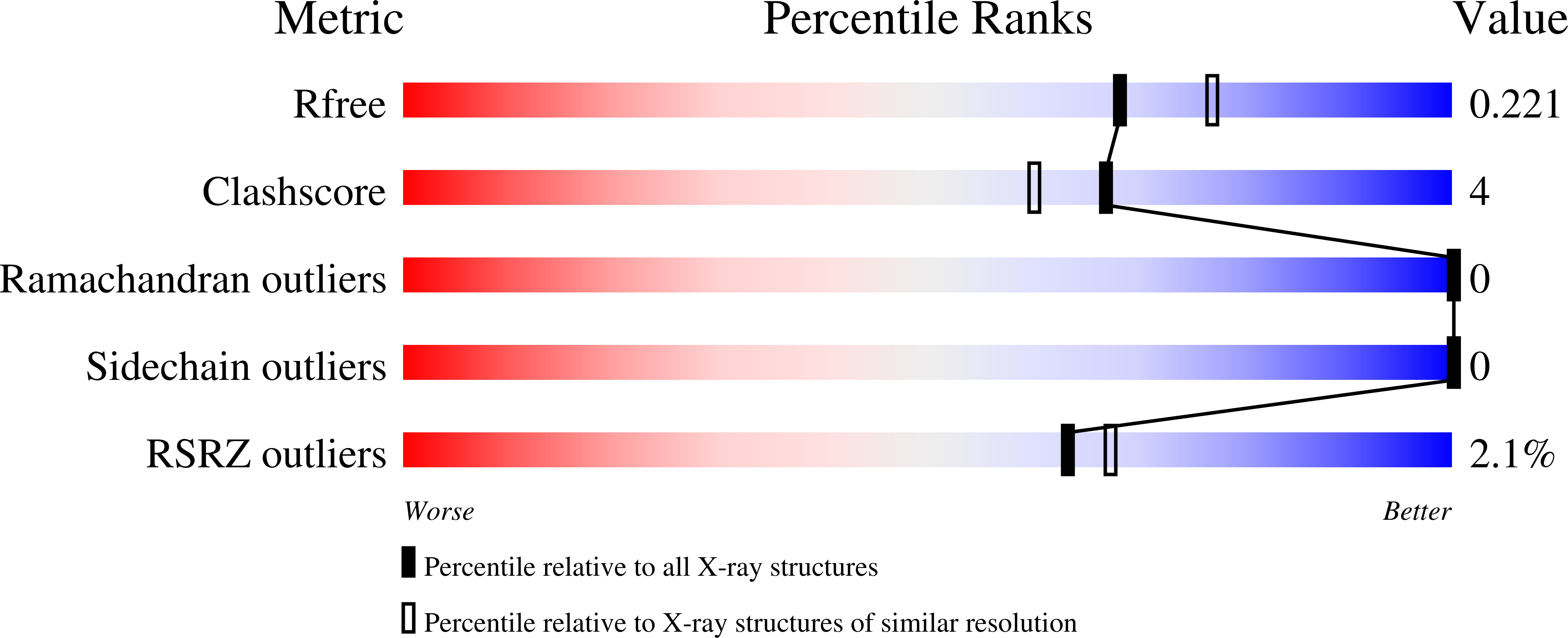
A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
Hadhazi, A., Li, L., Bailly, B., Maggioni, A., Martin, G., Dirr, L., Dyason, J.C., Thomson, R.J., Gao, G.F., Borbas, A., Ve, T., Pascolutti, M., von Itzstein, M.(2018) ChemMedChem 13: 785-789
- PubMed: 29453852
- DOI: https://doi.org/10.1002/cmdc.201800092
- Primary Citation of Related Structures:
6BR5, 6BR6 - PubMed Abstract:
Influenza virus infection continues to cause significant, often severe, respiratory illness worldwide. A validated target for the development of anti-influenza agents is the virus surface protein sialidase. In the current study, we have discovered a highly potent inhibitor of influenza virus sialidase, based on a novel sialosyl sulfonate template. The synthesised 3-guanidino sialosyl α-sulfonate, a sulfonozanamivir analogue, inhibits viral replication in vitro at the nanomolar level, comparable to that of the anti-influenza drug zanamivir. Using protein X-ray crystallography we show that the sialosyl α-sulfonate template binds within the sialidase active site in a 1 C 4 chair conformation. The C1-sulfonate moiety forms crucial and strong-binding interactions with the active site's triarginyl cluster, while the 3-guanidino moiety interacts significantly with conserved active site residues. This sulfonozanamivir analogue provides a new direction in anti-influenza virus drug development.
Organizational Affiliation:
Institute for Glycomics, Gold Coast Campus, Griffith University, Queensland, 4222, Australia.