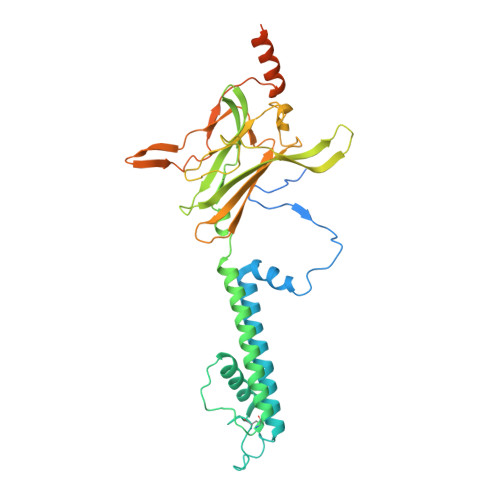
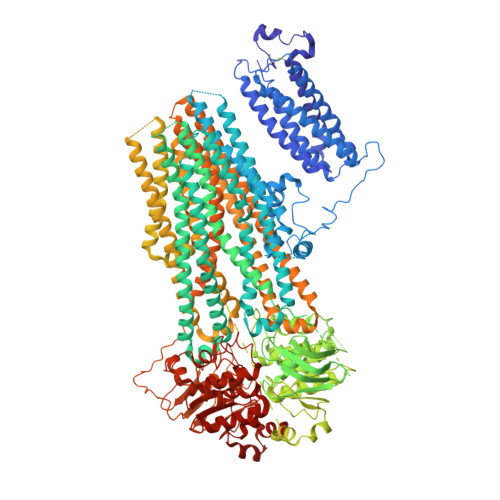
Molecular structure of human KATP in complex with ATP and ADP.
Lee, K.P.K., Chen, J., MacKinnon, R.(2017) Elife 6
- PubMed: 29286281
- DOI: https://doi.org/10.7554/eLife.32481
- Primary Citation of Related Structures:
6C3O, 6C3P - PubMed Abstract:
In many excitable cells, KATP channels respond to intracellular adenosine nucleotides: ATP inhibits while ADP activates. We present two structures of the human pancreatic KATP channel, containing the ABC transporter SUR1 and the inward-rectifier K + channel Kir6.2, in the presence of Mg 2+ and nucleotides. These structures, referred to as quatrefoil and propeller forms, were determined by single-particle cryo-EM at 3.9 Å and 5.6 Å, respectively. In both forms, ATP occupies the inhibitory site in Kir6.2. The nucleotide-binding domains of SUR1 are dimerized with Mg 2+ -ATP in the degenerate site and Mg 2+ -ADP in the consensus site. A lasso extension forms an interface between SUR1 and Kir6.2 adjacent to the ATP site in the propeller form and is disrupted in the quatrefoil form. These structures support the role of SUR1 as an ADP sensor and highlight the lasso extension as a key regulatory element in ADP's ability to override ATP inhibition.
- Laboratory of Molecular Neurobiology and Biophysics, Howard Hughes Medical Institute, The Rockefeller University, New York, United States.
Organizational Affiliation: