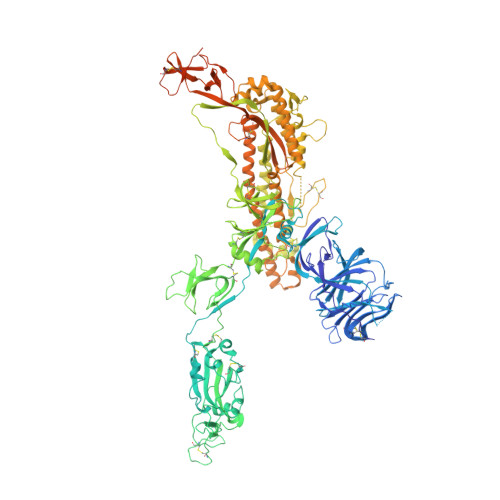
Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Kirchdoerfer, R.N., Wang, N., Pallesen, J., Wrapp, D., Turner, H.L., Cottrell, C.A., Corbett, K.S., Graham, B.S., McLellan, J.S., Ward, A.B.(2018) Sci Rep 8: 15701-15701
- PubMed: 30356097
- DOI: https://doi.org/10.1038/s41598-018-34171-7
- Primary Citation of Related Structures:
6CRV, 6CRW, 6CRX, 6CRZ, 6CS0, 6CS1, 6CS2 - PubMed Abstract:
Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 as a highly transmissible pathogenic human betacoronavirus. The viral spike glycoprotein (S) utilizes angiotensin-converting enzyme 2 (ACE2) as a host protein receptor and mediates fusion of the viral and host membranes, making S essential to viral entry into host cells and host species tropism. As SARS-CoV enters host cells, the viral S is believed to undergo a number of conformational transitions as it is cleaved by host proteases and binds to host receptors. We recently developed stabilizing mutations for coronavirus spikes that prevent the transition from the pre-fusion to post-fusion states. Here, we present cryo-EM analyses of a stabilized trimeric SARS-CoV S, as well as the trypsin-cleaved, stabilized S, and its interactions with ACE2. Neither binding to ACE2 nor cleavage by trypsin at the S1/S2 cleavage site impart large conformational changes within stabilized SARS-CoV S or expose the secondary cleavage site, S2'.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, 92037, USA.
Organizational Affiliation: