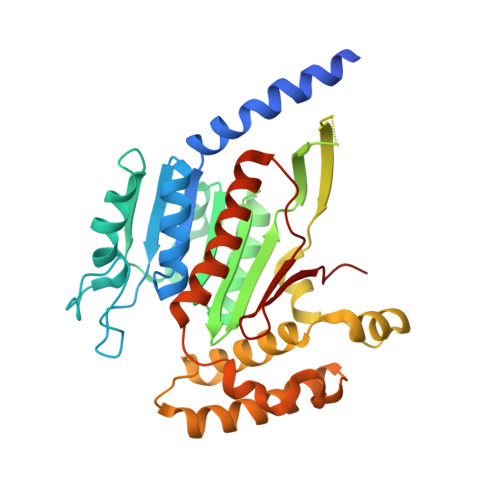
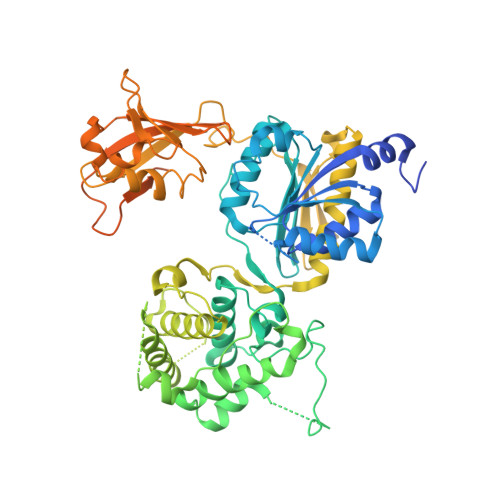
Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Lv, Z., Yuan, L., Atkison, J.H., Williams, K.M., Vega, R., Sessions, E.H., Divlianska, D.B., Davies, C., Chen, Y., Olsen, S.K.(2018) Nat Commun 9: 5145-5145
- PubMed: 30514846
- DOI: https://doi.org/10.1038/s41467-018-07015-1
- Primary Citation of Related Structures:
6CWY, 6CWZ - PubMed Abstract:
E1 enzymes activate ubiquitin (Ub) and ubiquitin-like modifiers (Ubls) in the first step of Ub/Ubl conjugation cascades and represent potential targets for therapeutic intervention in cancer and other life-threatening diseases. Here, we report the crystal structure of the E1 enzyme for the Ubl SUMO in complex with a recently discovered and highly specific covalent allosteric inhibitor (COH000). The structure reveals that COH000 targets a cryptic pocket distinct from the active site that is completely buried in all previous SUMO E1 structures and that COH000 binding to SUMO E1 is accompanied by a network of structural changes that altogether lock the enzyme in a previously unobserved inactive conformation. These structural changes include disassembly of the active site and a 180° rotation of the catalytic cysteine-containing SCCH domain, relative to conformational snapshots of SUMO E1 poised to catalyze adenylation. Altogether, our study provides a molecular basis for the inhibitory mechanism of COH000 and its SUMO E1 specificity, and also establishes a framework for potential development of molecules targeting E1 enzymes for other Ubls at a cryptic allosteric site.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology and Hollings Cancer Center, Medical University of South Carolina, Charleston, 29425, SC, USA.