De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
Thomas, F., Dawson, W.M., Lang, E.J.M., Burton, A.J., Bartlett, G.J., Rhys, G.G., Mulholland, A.J., Woolfson, D.N.(2018) ACS Synth Biol 7: 1808-1816
- PubMed: 29944338
- DOI: https://doi.org/10.1021/acssynbio.8b00225
- Primary Citation of Related Structures:
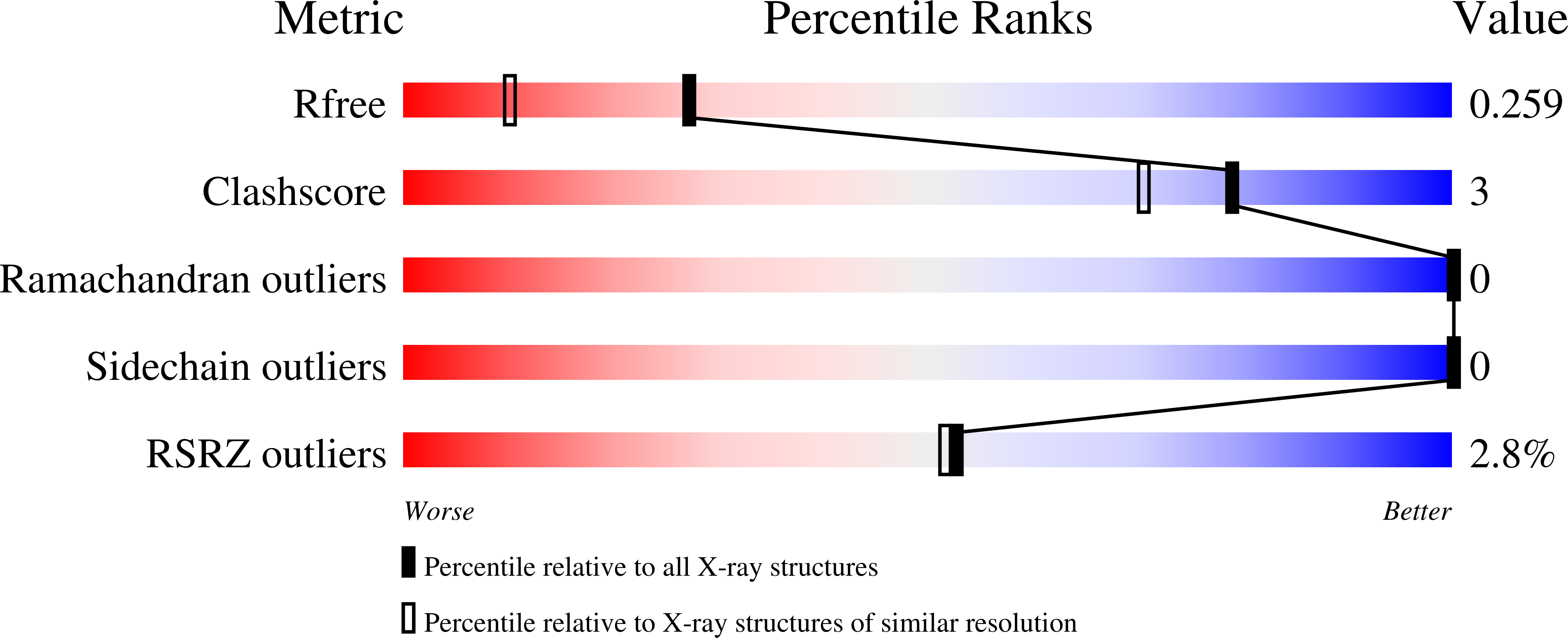
6EIK, 6EIZ - PubMed Abstract:
We describe de novo-designed α-helical barrels (αHBs) that bind and discriminate between lipophilic biologically active molecules. αHBs have five or more α-helices arranged around central hydrophobic channels the diameters of which scale with oligomer state. We show that pentameric, hexameric, and heptameric αHBs bind the environmentally sensitive dye 1,6-diphenylhexatriene (DPH) in the micromolar range and fluoresce. Displacement of the dye is used to report the binding of nonfluorescent molecules: palmitic acid and retinol bind to all three αHBs with submicromolar inhibitor constants; farnesol binds the hexamer and heptamer; but β-carotene binds only the heptamer. A co-crystal structure of the hexamer with farnesol reveals oriented binding in the center of the hydrophobic channel. Charged side chains engineered into the lumen of the heptamer facilitate binding of polar ligands: a glutamate variant binds a cationic variant of DPH, and introducing lysine allows binding of the biosynthetically important farnesol diphosphate.
Organizational Affiliation:
School of Chemistry , University of Bristol , Cantock's Close , Bristol BS8 1TS , U.K.