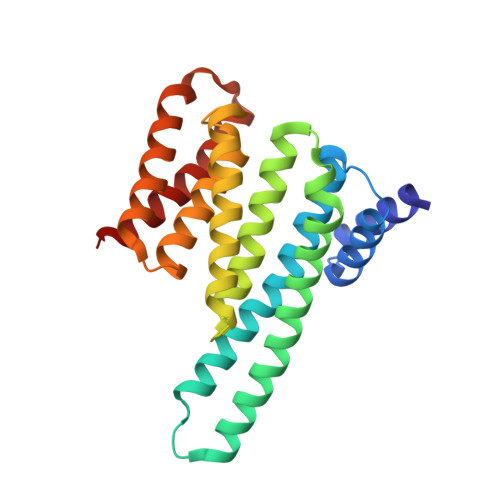
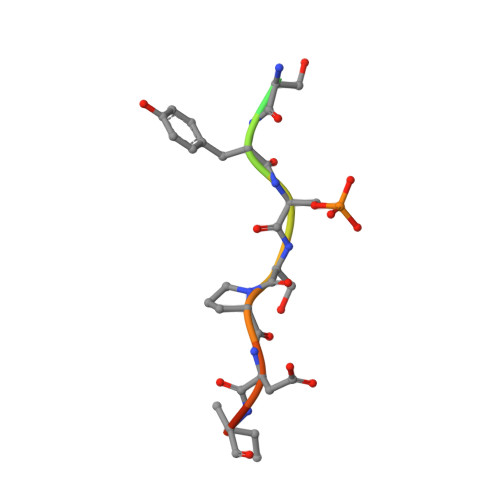
Biophysical and structural insight into the USP8/14-3-3 interaction.
Centorrino, F., Ballone, A., Wolter, M., Ottmann, C.(2018) FEBS Lett 592: 1211-1220
- PubMed: 29473952
- DOI: https://doi.org/10.1002/1873-3468.13017
- Primary Citation of Related Structures:
6F09 - PubMed Abstract:
The ubiquitin-specific protease 8 (USP8)/14-3-3 protein-protein interaction has recently been shown to exert a significant role in the pathogenesis of Cushing's disease (CD). USP8 is a deubiquitinase that prevents epidermal growth factor receptor (EGFR) degradation. Impairment of 14-3-3 binding leads to a higher deubiquitination of EGFR and results in a higher EGFR signaling and an increased production of adrenocorticotropic hormone. Here we report the high-resolution crystal structure of the 14-3-3 binding motif of USP8 surrounding Ser718 in complex with 14-3-3ζ and characterize the interaction with fluorescence polarization and isothermal titration calorimetry. Furthermore, we analyze the effect of USP8 mutations identified in CD on binding to 14-3-3.
- Laboratory of Chemical Biology, Department of Biomedical Engineering, Institute for Complex Molecular Systems, Eindhoven University of Technology, The Netherlands.
Organizational Affiliation: