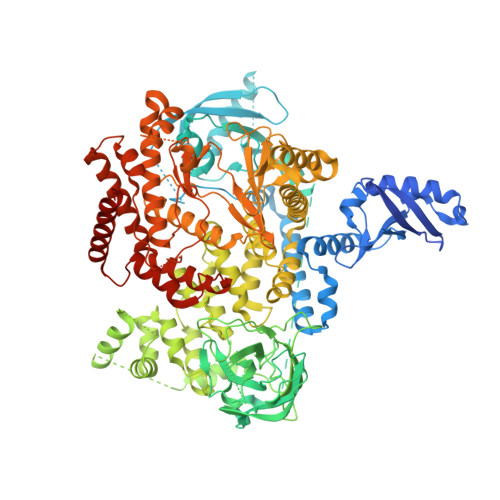
Discovery of a Novel Inhaled PI3K delta Inhibitor for the Treatment of Respiratory Diseases.
Erra, M., Taltavull, J., Bernal, F.J., Caturla, J.F., Carrascal, M., Pages, L., Mir, M., Espinosa, S., Gracia, J., Dominguez, M., Sabate, M., Paris, S., Maldonado, M., Hernandez, B., Bravo, M., Calama, E., Miralpeix, M., Lehner, M.D., Calbet, M.(2018) J Med Chem 61: 9551-9567
- PubMed: 30351000
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00873
- Primary Citation of Related Structures:
6G6W - PubMed Abstract:
Oral PI3Kδ inhibitors such as Idelalisib and Duvelisib have shown efficacy as anticancer agents and Idelalisib has been approved for the treatment of three B-cell cancers. However, Idelalisib has a black box warning on its product label regarding the risks of fatal and serious toxicities including hepatic toxicity, severe diarrhea, colitis, pneumonitis, infections, and intestinal perforation. Some of these side effects are mechanism-related and could hinder the development of Idelalisib for less severe conditions. For respiratory diseases, compounds administered by inhalation are delivered directly to the site of action and may improve the therapeutic index of a drug, minimizing undesired side effects. This work describes the discovery and optimization of inhaled PI3Kδ inhibitors intended for the treatment of severe asthma and COPD. Once the potency was in the desired range, efforts were focused on identifying the particular physicochemical properties that could translate into better lung retention. This medicinal chemistry exercise led to the identification of LAS195319 as a candidate for clinical development.