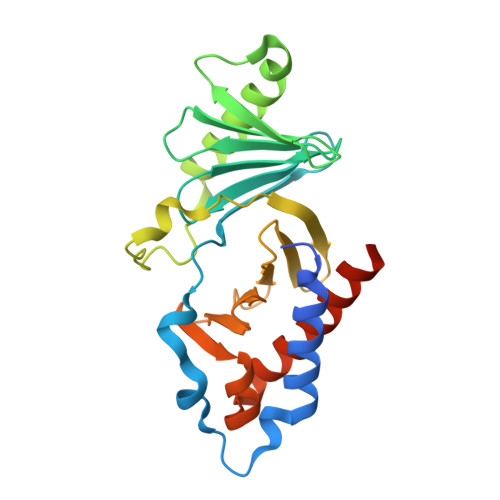
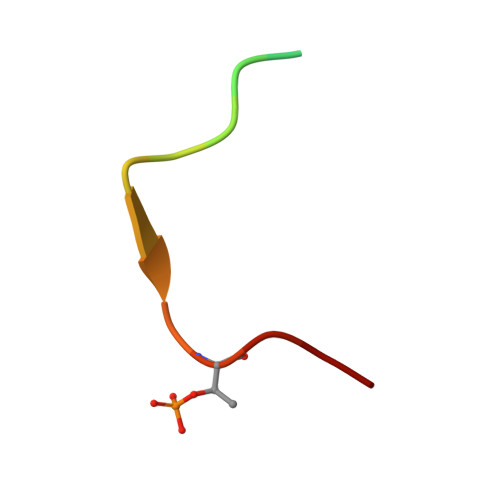
Proper chromosome alignment depends on BRCA2 phosphorylation by PLK1.
Ehlen, A., Martin, C., Miron, S., Julien, M., Theillet, F.X., Ropars, V., Sessa, G., Beaurepere, R., Boucherit, V., Duchambon, P., El Marjou, A., Zinn-Justin, S., Carreira, A.(2020) Nat Commun 11: 1819-1819
- PubMed: 32286328
- DOI: https://doi.org/10.1038/s41467-020-15689-9
- Primary Citation of Related Structures:
6GY2 - PubMed Abstract:
The BRCA2 tumor suppressor protein is involved in the maintenance of genome integrity through its role in homologous recombination. In mitosis, BRCA2 is phosphorylated by Polo-like kinase 1 (PLK1). Here we describe how this phosphorylation contributes to the control of mitosis. We identify a conserved phosphorylation site at T207 of BRCA2 that constitutes a bona fide docking site for PLK1 and is phosphorylated in mitotic cells. We show that BRCA2 bound to PLK1 forms a complex with the phosphatase PP2A and phosphorylated-BUBR1. Reducing BRCA2 binding to PLK1, as observed in BRCA2 breast cancer variants S206C and T207A, alters the tetrameric complex resulting in unstable kinetochore-microtubule interactions, misaligned chromosomes, faulty chromosome segregation and aneuploidy. We thus reveal a role of BRCA2 in the alignment of chromosomes, distinct from its DNA repair function, with important consequences on chromosome stability. These findings may explain in part the aneuploidy observed in BRCA2-mutated tumors.
- Institut Curie, PSL Research University, CNRS, UMR3348, F-91405, Orsay, France.
Organizational Affiliation: