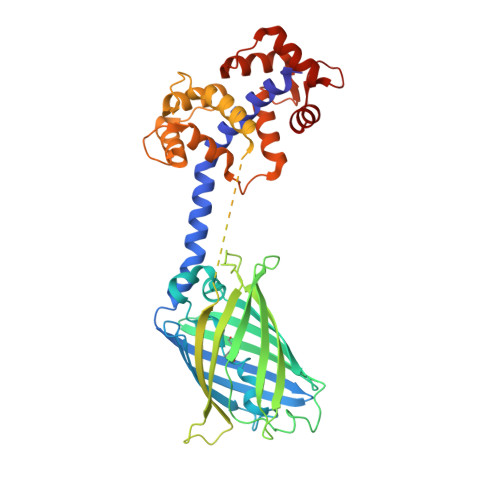
Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Collu, G., Bierig, T., Krebs, A.S., Engilberge, S., Varma, N., Guixa-Gonzalez, R., Sharpe, T., Deupi, X., Olieric, V., Poghosyan, E., Benoit, R.M.(2021) Structure
- PubMed: 34587504
- DOI: https://doi.org/10.1016/j.str.2021.09.002
- Primary Citation of Related Structures:
6HR1, 6XYR, 6YT3 - PubMed Abstract:
Chimeric fusion proteins are essential tools for protein nanotechnology. Non-optimized protein-protein connections are usually flexible and therefore unsuitable as structural building blocks. Here we show that the ER/K motif, a single α-helical domain (SAH), can be seamlessly fused to terminal helices of proteins, forming an extended, partially free-standing rigid helix. This enables the connection of two domains at a defined distance and orientation. We designed three constructs termed YFPnano, T4Lnano, and MoStoNano. Analysis of experimentally determined structures and molecular dynamics simulations reveals a certain degree of plasticity in the connections that allows the adaptation to crystal contact opportunities. Our data show that SAHs can be stably integrated into designed structural elements, enabling new possibilities for protein nanotechnology, for example, to improve the exposure of epitopes on nanoparticles (structural vaccinology), to engineer crystal contacts with minimal impact on construct flexibility (for the study of protein dynamics), and to design novel biomaterials.
- Laboratory of Nanoscale Biology, Division of Biology and Chemistry, Paul Scherrer Institute, 5232 Villigen PSI, Switzerland; Department of Biology, ETH Zürich, 8093 Zürich, Switzerland.
Organizational Affiliation: